8B7O
 
 | |
8B7P
 
 | | Crystal structure of an AA9 LPMO from Aspergillus nidulans, AnLPMOC | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, Endo-beta-1,4-glucanase D | | Authors: | Males, A, Rafael Fanchini Terrasan, C, Davies, G.J, Walton, P.H. | | Deposit date: | 2022-09-30 | | Release date: | 2023-10-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Characterisation of lytic polysaccharide monooxygenases from Aspergillus nidulans
To Be Published
|
|
8B7Y
 
 | | Cryo-EM structure of the E.coli 70S ribosome in complex with the antibiotic Myxovalargin B. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Koller, T.O, Graf, M, Wilson, D.N. | | Deposit date: | 2022-10-03 | | Release date: | 2023-01-25 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The Myxobacterial Antibiotic Myxovalargin: Biosynthesis, Structural Revision, Total Synthesis, and Molecular Characterization of Ribosomal Inhibition.
J.Am.Chem.Soc., 145, 2023
|
|
8B9A
 
 | |
8B9B
 
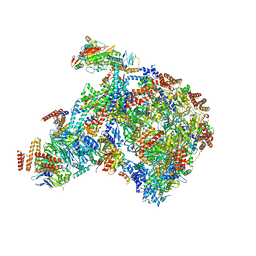 | |
8B9C
 
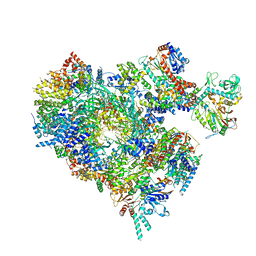 | | S. cerevisiae pol alpha - replisome complex | | Descriptor: | Cell division control protein 45, Chromosome segregation in meiosis protein 3, DNA polymerase alpha catalytic subunit A, ... | | Authors: | Jones, M.L, Yeeles, J.T.P. | | Deposit date: | 2022-10-05 | | Release date: | 2023-08-09 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | How Pol alpha-primase is targeted to replisomes to prime eukaryotic DNA replication.
Mol.Cell, 83, 2023
|
|
8B9D
 
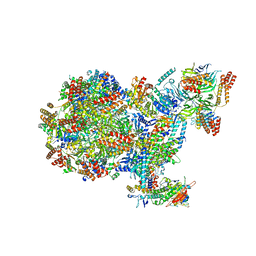 | | Human replisome bound by Pol Alpha | | Descriptor: | Cell division control protein 45 homolog, Claspin, DNA Molecule, ... | | Authors: | Jones, M.L, Yeeles, J.T.P. | | Deposit date: | 2022-10-05 | | Release date: | 2023-08-09 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | How Pol alpha-primase is targeted to replisomes to prime eukaryotic DNA replication.
Mol.Cell, 83, 2023
|
|
8BA7
 
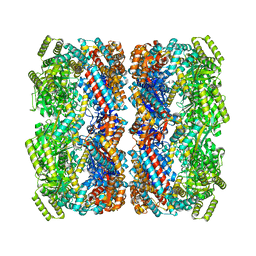 | |
8BA8
 
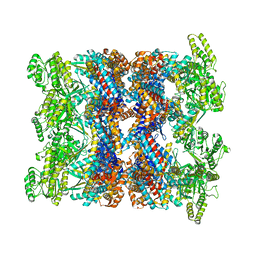 | | CryoEM structure of GroEL-ADP.BeF3-Rubisco. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chaperonin GroEL, ... | | Authors: | Gardner, S, Saibil, H.R. | | Deposit date: | 2022-10-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of substrate progression through the bacterial chaperonin cycle.
Proc Natl Acad Sci U S A, 120, 2023
|
|
8BA9
 
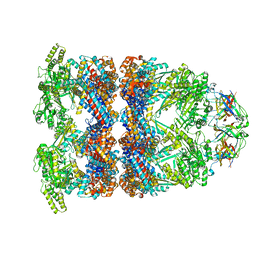 | | CryoEM structure of GroEL-GroES-ADP.AlF3-Rubisco. | | Descriptor: | 60 kDa chaperonin, ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, ... | | Authors: | Gardner, S, Saibil, H.R. | | Deposit date: | 2022-10-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of substrate progression through the bacterial chaperonin cycle.
Proc Natl Acad Sci U S A, 120, 2023
|
|
8BAC
 
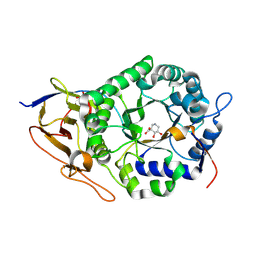 | | Crystal structure of human heparanase in complex with competitive inhibitor GD05 | | Descriptor: | (3S,4R,5R)-4,5-dihydroxypiperidine-3-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Armstrong, Z, Davies, G.J. | | Deposit date: | 2022-10-11 | | Release date: | 2023-03-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Synthesis of Uronic Acid 1-Azasugars as Putative Inhibitors of alpha-Iduronidase, beta-Glucuronidase and Heparanase.
Chembiochem, 24, 2023
|
|
8BBZ
 
 | |
8BCP
 
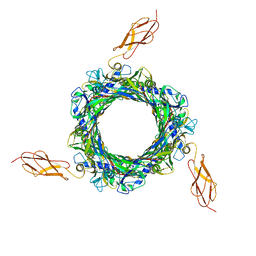 | |
8BCU
 
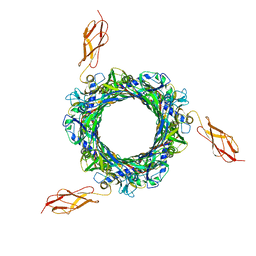 | |
8BD3
 
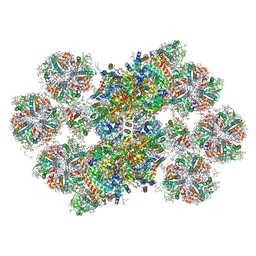 | | Cryo-EM structure of the Photosystem II - LHCII supercomplex from Chlorella ohadi | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R)-beta,beta-caroten-3-ol, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Fadeeva, M, Klaiman, D, Caspy, I, Nelson, N. | | Deposit date: | 2022-10-18 | | Release date: | 2023-08-09 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Structure of Chlorella ohadii Photosystem II Reveals Protective Mechanisms against Environmental Stress.
Cells, 12, 2023
|
|
8BD6
 
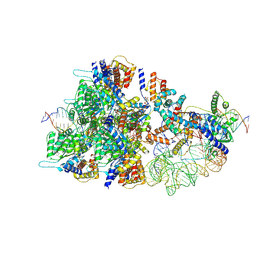 | | Cas12k-sgRNA-dsDNA-TnsC non-productive complex. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cas12k, DNA, ... | | Authors: | Schmitz, M, Querques, I, Oberli, S, Chanez, C, Jinek, M. | | Deposit date: | 2022-10-18 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis for the assembly of the type V CRISPR-associated transposon complex.
Cell, 185, 2022
|
|
8BD7
 
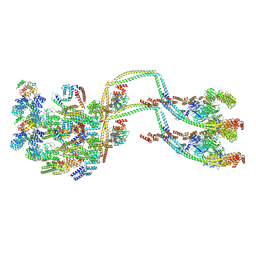 | | IFTB1 subcomplex of anterograde Intraflagellar transport trains (Chlamydomonas reinhardtii) | | Descriptor: | Clusterin-associated protein 1, IFT54, IFT70, ... | | Authors: | Lacey, S.E, Foster, H.E, Pigino, G. | | Deposit date: | 2022-10-18 | | Release date: | 2023-01-11 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (9.9 Å) | | Cite: | The molecular structure of IFT-A and IFT-B in anterograde intraflagellar transport trains.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8BDA
 
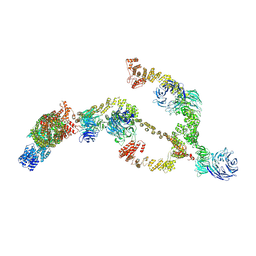 | | IFTA complex in anterograde intraflagellar transport trains (Chlamydomonas reinhardtii) | | Descriptor: | Intraflagellar transport particle protein 140, Intraflagellar transport protein 121, Intraflagellar transport protein 122 homolog, ... | | Authors: | Lacey, S.E, Foster, H.E, Pigino, G. | | Deposit date: | 2022-10-18 | | Release date: | 2023-01-11 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (20.7 Å) | | Cite: | The molecular structure of IFT-A and IFT-B in anterograde intraflagellar transport trains.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8BE3
 
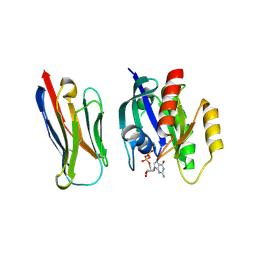 | | Crystal structure of KRasG12V-Nanobody84 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, MAGNESIUM ION, ... | | Authors: | Fischer, B, Wohlkonig, A, Steyaert, J. | | Deposit date: | 2022-10-21 | | Release date: | 2023-11-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Allosteric nanobodies to study the interactions between SOS1 and RAS.
Nat Commun, 15, 2024
|
|
8BE4
 
 | |
8BE5
 
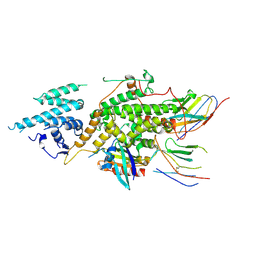 | | Crystal structure of SOS1-KRasG12V-Nanobody22-Nanobody75 | | Descriptor: | Isoform 2B of GTPase KRas, Nanobody22, Nanobody75, ... | | Authors: | Fischer, B, Wohlkonig, A, Steyaert, J. | | Deposit date: | 2022-10-21 | | Release date: | 2023-11-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | Allosteric nanobodies to study the interactions between SOS1 and RAS.
Nat Commun, 15, 2024
|
|
8BF7
 
 | |
8BFL
 
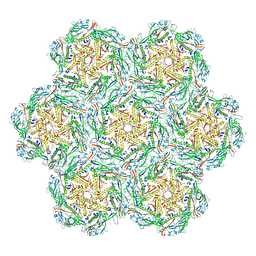 | | Jumbo Phage phi-kp24 empty capsid hexamers | | Descriptor: | Major head protein | | Authors: | Ouyang, R. | | Deposit date: | 2022-10-26 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | High-resolution reconstruction of a Jumbo-bacteriophage infecting capsulated bacteria using hyperbranched tail fibers.
Nat Commun, 13, 2022
|
|
8BFP
 
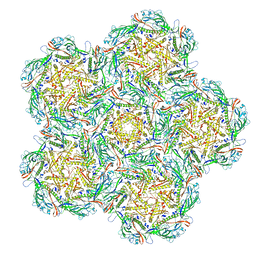 | | Jumbo Phage phi-kp24 empty capsid pentamer hexamers | | Descriptor: | Major head protein | | Authors: | Ouyang, R. | | Deposit date: | 2022-10-26 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | High-resolution reconstruction of a Jumbo-bacteriophage infecting capsulated bacteria using hyperbranched tail fibers.
Nat Commun, 13, 2022
|
|
8BGB
 
 | |






