9FSV
 
 | | Yeast 20S proteasome with human beta2i (1-53) in complex with epoxyketone inhibitor 42 | | Descriptor: | (2S)-N-[(2S)-1-[[(1S)-2-cyclohexyl-1-[(2R,3S,6R,7S)-3-methanoyl-2,6-dimethyl-6,7-bis(oxidanyl)-1,4-oxazepan-7-yl]ethyl]amino]-3-(4-methoxyphenyl)-1-oxidanylidene-propan-2-yl]-2-(2-morpholin-4-ylethanoylamino)-4-oxidanyl-butanamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Maurits, E, Huber, E.M, Dekker, P.M, Wang, X, Heinemeyer, W, Florea, B.I, Groll, M, Overkleeft, H.S. | | Deposit date: | 2024-06-22 | | Release date: | 2024-07-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure-based design of peptide epoxyketones selectively targeting the three human immunoproteasome active sites
to be published
|
|
9FT0
 
 | | Yeast 20S proteasome in complex with epoxyketone inhibitor 42 | | Descriptor: | (2S)-N-[(2S)-1-[[(1S)-2-cyclohexyl-1-[(2R,3S,6R,7S)-3-methanoyl-2,6-dimethyl-6,7-bis(oxidanyl)-1,4-oxazepan-7-yl]ethyl]amino]-3-(4-methoxyphenyl)-1-oxidanylidene-propan-2-yl]-2-(2-morpholin-4-ylethanoylamino)-4-oxidanyl-butanamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Maurits, E, Huber, E.M, Dekker, P.M, Wang, X, Heinemeyer, W, Florea, B.I, Groll, M, Overkleeft, H.S. | | Deposit date: | 2024-06-23 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure-based design of peptide epoxyketones selectively targeting the three human immunoproteasome active sites
to be published
|
|
9FT1
 
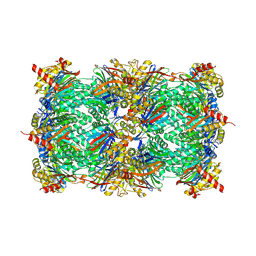 | | Yeast 20S proteasome in complex with epoxyketone inhibitor 13 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, Probable proteasome subunit alpha type-7, ... | | Authors: | Maurits, E, Huber, E.M, Dekker, P.M, Wang, X, Heinemeyer, W, Florea, B.I, Groll, M, Overkleeft, H.S. | | Deposit date: | 2024-06-23 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based design of peptide epoxyketones selectively targeting the three human immunoproteasome active sites
to be published
|
|
9FTC
 
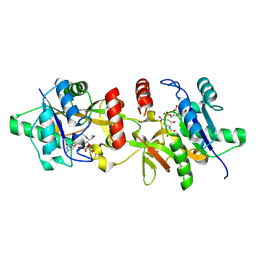 | | Aeromonas caviae CMP-Pse5Ac7Ac synthetase in the presence of CMP-Pse5Ac7Ac. | | Descriptor: | (2~{R},4~{S},5~{S},6~{S})-5-acetamido-6-[(1~{S},2~{S})-1-acetamido-2-oxidanyl-propyl]-2-[[(2~{R},3~{S},4~{R},5~{R})-5-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-4-oxidanyl-oxane-2-carboxylic acid, NeuA | | Authors: | Cowan, A.R, Hemsworth, G.R, Keenan, T, Fascione, M.A. | | Deposit date: | 2024-06-24 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into a CMP-pseudaminic acid synthetase (PseF)
Structure, 2024
|
|
9FTQ
 
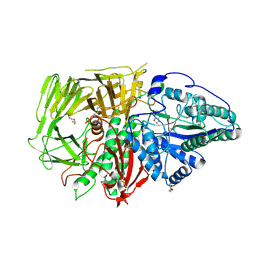 | | Drosophila golgi alpha-mannosidase II (dGMII) in complex with swainsonine-configured alkyl indolizidine | | Descriptor: | Alpha-mannosidase 2, DI(HYDROXYETHYL)ETHER, ZINC ION, ... | | Authors: | Bennett, M, Koemans, T, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2024-06-25 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structure-guided design of C3-branched swainsonine as potent and selective human Golgi alpha-mannosidase (GMII) inhibitor.
Chem.Commun.(Camb.), 2024
|
|
9FTR
 
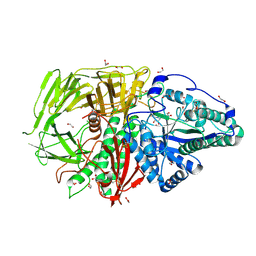 | | Drosophila golgi alpha-mannosidase II (dGMII) in complex with amide modified swainsonine-configured alkyl indolizidine | | Descriptor: | 1,2-ETHANEDIOL, Alpha-mannosidase 2, ZINC ION, ... | | Authors: | Bennett, M, Koemans, T, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2024-06-25 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structure-guided design of C3-branched swainsonine as potent and selective human Golgi alpha-mannosidase (GMII) inhibitor.
Chem.Commun.(Camb.), 2024
|
|
9FWG
 
 | | LSD1/CoREST bound to bomedemstat | | Descriptor: | Bomedemstat FAD adduct, Lysine-specific histone demethylase 1A, REST corepressor 1 | | Authors: | Speranzini, V, Mattevi, A. | | Deposit date: | 2024-06-30 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Characterization of structural, biochemical, pharmacokinetic, and pharmacodynamic properties of the LSD1 inhibitor bomedemstat in preclinical models.
Prostate, 84, 2024
|
|
9FWX
 
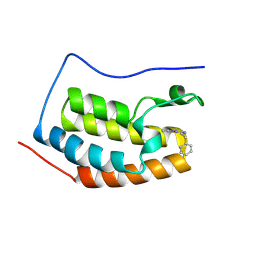 | | Crystal structure of BRD4 BD1 with MEN1404BS. | | Descriptor: | 3-methyl-~{N}-(2-phenylethyl)-[1,2,4]triazolo[4,3-b]pyridazin-6-amine, Bromodomain-containing protein 4 | | Authors: | Bader, G, Kessler, D. | | Deposit date: | 2024-07-01 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Probing Protein-Ligand Methyl-pi Interaction Geometries through Chemical Shift Measurements of Selectively Labeled Methyl Groups.
J.Med.Chem., 67, 2024
|
|
9FYD
 
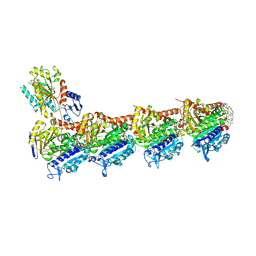 | | tubulin - cryptophycin-uD[Dab] complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Dessin, C, Schachtsiek, T, Voss, J, Abel, A.-C, Neumann, B, Stammler, H.-G, Prota, A.E, Sewald, N. | | Deposit date: | 2024-07-03 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Highly Cytotoxic Cryptophycin Derivatives with Modification in Unit D for Conjugation.
Angew.Chem.Int.Ed.Engl., 2024
|
|
9FYF
 
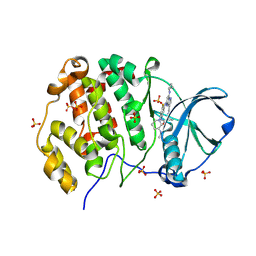 | | Crystal structure of human Casein Kinase II subunit alpha (CK2a1) in complex with TR06772818 | | Descriptor: | Casein kinase II subunit alpha, SULFATE ION, ~{N}-[5-[[3-cyano-7-(cyclopropylamino)pyrazolo[1,5-a]pyrimidin-5-yl]amino]-4-fluoranyl-2-[(3~{S})-3-(methylamino)piperidin-1-yl]phenyl]propanamide | | Authors: | Kraemer, A, Ong, H.W, Yang, X, Brown, J.W, Chang, E, Willson, T, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-07-03 | | Release date: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of human Casein Kinase II subunit alpha (CK2a1) in complex with TR06772818
To Be Published
|
|
9G13
 
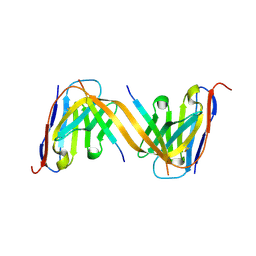 | | VHH H3-2 in complex with Tau C-terminal peptide | | Descriptor: | Isoform Tau-F of Microtubule-associated protein tau, VHH H3-2 | | Authors: | Dupre, E, Landrieu, I, Danis, C, Hanoulle, X, Mortelecque, J. | | Deposit date: | 2024-07-09 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | VHH H3-2 in complex with Tau C-terminal peptide
To Be Published
|
|
9G3X
 
 | |
9G3Y
 
 | |
9G3Z
 
 | |
9G7H
 
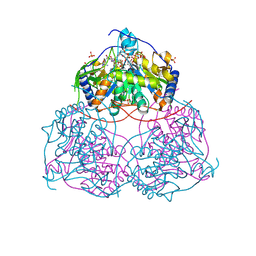 | | Human Sirt6 in complex with ADP-ribose and the inhibitor 2-Pr | | Descriptor: | 2,4-bis(oxidanylidene)-1-[2-oxidanylidene-2-[[(2S)-3-oxidanylidene-3-(propylamino)-2-[3-(3-pyridin-3-yl-1,2,4-oxadiazol-5-yl)propanoylamino]propyl]amino]ethyl]pyrimidine-5-carboxamide, NAD-dependent protein deacetylase sirtuin-6, SULFATE ION, ... | | Authors: | You, W, Steegborn, C. | | Deposit date: | 2024-07-21 | | Release date: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Elucidating the Unconventional Binding Mode of a DNA-Encoded Library Hit Provides a Blueprint for Sirtuin 6 Inhibitor Development.
Chemmedchem, 2024
|
|
9GAO
 
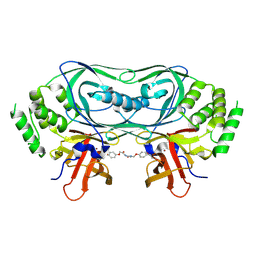 | | Crystal structure of CRBNmidi in complex with 2-(4-(2,6-dioxopiperidin-3-yl)phenoxy)-N-methylacetamide | | Descriptor: | 2-[4-[(3~{R})-2,6-bis(oxidanylidene)piperidin-3-yl]phenoxy]-~{N}-methyl-ethanamide, Protein cereblon, ZINC ION | | Authors: | Rutter, Z.J, Kroupova, A, Zollman, D, Ciulli, A. | | Deposit date: | 2024-07-29 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design of a Cereblon construct for crystallographic and biophysical studies of protein degraders
To Be Published
|
|
9GBE
 
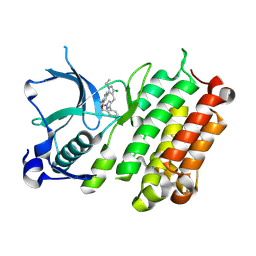 | |
9GD1
 
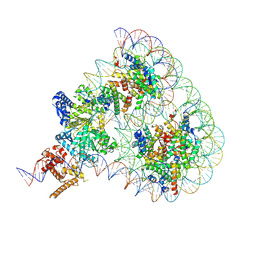 | | Structure of Chd1 bound to a hexasome-nucleosome complex with a dyad-to-dyad distance of 103 bp. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromo domain-containing protein 1, ... | | Authors: | Engeholm, M, Roske, J.J, Oberbeckmann, E, Dienemann, C, Lidschreiber, M, Cramer, P, Farnung, L. | | Deposit date: | 2024-08-04 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Resolution of transcription-induced hexasome-nucleosome complexes by Chd1 and FACT.
Mol.Cell, 84, 2024
|
|
9GD2
 
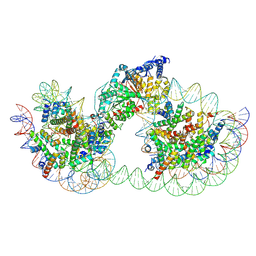 | | Structure of Chd1 bound to a dinucleosome with a dyad-to-dyad distance of 103 bp. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromo domain-containing protein 1, ... | | Authors: | Engeholm, M, Roske, J.J, Oberbeckmann, E, Dienemann, C, Lidschreiber, M, Cramer, P, Farnung, L. | | Deposit date: | 2024-08-04 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Resolution of transcription-induced hexasome-nucleosome complexes by Chd1 and FACT.
Mol.Cell, 84, 2024
|
|
9GMO
 
 | |
9GRP
 
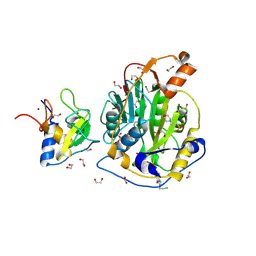 | | SARS-CoV-2 methyltransferase nsp10-16 in complex with SAM and beta-chloroethyl theophylline | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D, Kiene, A. | | Deposit date: | 2024-09-12 | | Release date: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of SARS-CoV-2 methyltransferase nsp10-16 with Cap0-site binders
To Be Published
|
|
9GTF
 
 | | SARS-CoV-2 methyltransferase nsp10-16 in complex with SAM and theophylline derivative LAS 57256190 | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D, Kiene, A. | | Deposit date: | 2024-09-17 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structures of SARS-CoV-2 methyltransferase nsp10-16 with Cap0-site binders
To Be Published
|
|
9GUE
 
 | | SARS-CoV-2 methyltransferase nsp10-16 in complex with SAM and theophylline derivative LAS 57256189 | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D, Kiene, A. | | Deposit date: | 2024-09-19 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of SARS-CoV-2 methyltransferase nsp10-16 with Cap0-site binders
To Be Published
|
|
9GUF
 
 | | SARS-CoV-2 methyltransferase nsp10-16 in complex with SAM and theophylline derivative LAS 54571106 | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D, Kiene, A. | | Deposit date: | 2024-09-19 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of SARS-CoV-2 methyltransferase nsp10-16 with Cap0-site binders
To Be Published
|
|
9IIW
 
 | |






