9D9K
 
 | |
9EMA
 
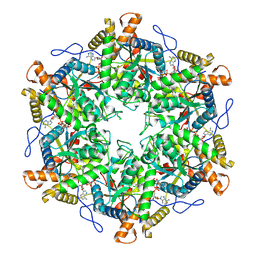 | | RUVBL1/2 in complex with ATP and CB-6644 inhibitor | | Descriptor: | 5-chloranyl-2-ethoxy-4-fluoranyl-~{N}-[4-[[3-(methoxymethyl)-1-oxidanylidene-6,7-dihydro-5~{H}-pyrazolo[1,2-a][1,2]benzodiazepin-2-yl]amino]-2,2-dimethyl-4-oxidanylidene-butyl]benzamide, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Lopez-Perrote, A, Llorca, O, Garcia-Martin, C. | | Deposit date: | 2024-03-07 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Mechanism of allosteric inhibition of RUVBL1-RUVBL2 by the small-molecule CB-6644
Cell Rep Phys Sci, 2024
|
|
9EN6
 
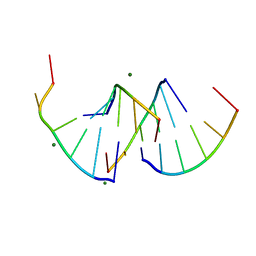 | | Crystal structure of RNA G2C4 repeats - native model pH 6.5 | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*GP*GP*CP*CP*CP*C)-3') | | Authors: | Mateja-Pluta, M, Kiliszek, A. | | Deposit date: | 2024-03-12 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (0.918 Å) | | Cite: | Antisense RNA C9orf72 hexanucleotide repeat associated with amyotrophic lateral sclerosis and frontotemporal dementia forms a triplex-like structure and binds small synthetic ligand.
Nucleic Acids Res., 52, 2024
|
|
9ENZ
 
 | |
9EO0
 
 | | Small-Molecule Inhibitors of Programmed Cell Death-1/Programmed Death-Ligand 1 | | Descriptor: | Programmed cell death 1 ligand 1, SULFATE ION, ~{N}-[3-[3-[[5-[(2-hydroxyethylamino)methyl]pyridin-2-yl]carbonylamino]-2-methyl-phenyl]-2-methyl-phenyl]-5-[[3-(methylsulfonylamino)propylamino]methyl]pyridine-2-carboxamide | | Authors: | Plewka, J, Hec, A, Sitar, T, Holak, T. | | Deposit date: | 2024-03-14 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Nonsymmetrically Substituted 1,1'-Biphenyl-Based Small Molecule Inhibitors of the PD-1/PD-L1 Interaction.
Acs Med.Chem.Lett., 15, 2024
|
|
9EO2
 
 | |
9EO5
 
 | |
9EO6
 
 | | SARS-CoV2 major protease in complex with a covalent inhibitor SLL11. | | Descriptor: | 3C-like proteinase nsp5, Inhibitor SLL11, POTASSIUM ION | | Authors: | Moche, M, Lennerstrand, J, Nyman, T, Strandback, E, Akaberi, D. | | Deposit date: | 2024-03-14 | | Release date: | 2024-09-04 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Identification of novel and potent inhibitors of SARS-CoV-2 main protease from DNA-encoded chemical libraries.
Antimicrob.Agents Chemother., 2024
|
|
9EO8
 
 | |
9EOJ
 
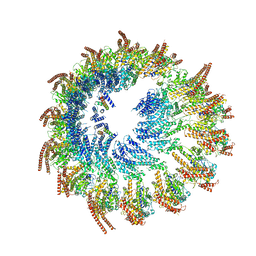 | | Vertebrate microtubule-capping gamma-tubulin ring complex | | Descriptor: | Gamma-tubulin complex component, Gamma-tubulin complex component 3 homolog, Gamma-tubulin complex component 6, ... | | Authors: | Vermeulen, B.J.A, Pfeffer, S. | | Deposit date: | 2024-03-15 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | gamma-TuRC asymmetry induces local protofilament mismatch at the RanGTP-stimulated microtubule minus end.
Embo J., 43, 2024
|
|
9EOR
 
 | | SARS-CoV2 major protease in complex with a covalent inhibitor SLL12. | | Descriptor: | 3C-like proteinase nsp5, Inhibitor SLL12, POTASSIUM ION | | Authors: | Moche, M, Lennerstrand, J, Nyman, T, Strandback, E, Akaberi, D. | | Deposit date: | 2024-03-15 | | Release date: | 2024-09-04 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Identification of novel and potent inhibitors of SARS-CoV-2 main protease from DNA-encoded chemical libraries.
Antimicrob.Agents Chemother., 2024
|
|
9EOU
 
 | |
9EOX
 
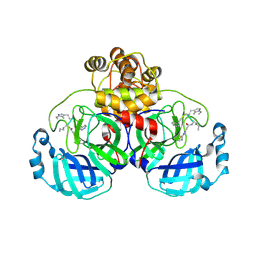 | | SARS-CoV2 major protease in covalent complex with a soluble inhibitor. | | Descriptor: | 3C-like proteinase nsp5, POTASSIUM ION, Soluble inhibitor | | Authors: | Moche, M, Lennerstrand, J, Nyman, T, Strandback, E, Akaberi, D. | | Deposit date: | 2024-03-15 | | Release date: | 2024-09-04 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Identification of novel and potent inhibitors of SARS-CoV-2 main protease from DNA-encoded chemical libraries.
Antimicrob.Agents Chemother., 2024
|
|
9EPV
 
 | | Crystal structure of human Casein Kinase II subunit alpha (CK2a1) in complex with allosteric ligand FGC333 | | Descriptor: | 1,2-ETHANEDIOL, 5-[[2-[(3-chloranyl-4-phenyl-phenyl)methylamino]-7-azaspiro[3.5]nonan-7-yl]sulfonyl]-1,3-dimethyl-benzimidazol-2-one, Casein kinase II subunit alpha, ... | | Authors: | Kraemer, A, Greco, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-03-20 | | Release date: | 2024-05-01 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Synthesis and evaluation of chemical linchpins for highly selective CK2 alpha targeting.
Eur.J.Med.Chem., 276, 2024
|
|
9EPW
 
 | | Crystal structure of human Casein Kinase II subunit alpha (CK2a1) in complex with allosteric ligand FGC3336 | | Descriptor: | Casein kinase II subunit alpha, SULFATE ION, ~{N}-[(3-chloranyl-4-phenyl-phenyl)methyl]-2-[1-(3-methylquinolin-8-yl)sulfonylpiperidin-4-yl]ethanamine | | Authors: | Kraemer, A, Greco, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-03-20 | | Release date: | 2024-05-01 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Synthesis and evaluation of chemical linchpins for highly selective CK2 alpha targeting.
Eur.J.Med.Chem., 276, 2024
|
|
9EPX
 
 | | Crystal structure of human Casein Kinase II subunit alpha (CK2a1) in complex with allosteric ligand FGC3331 | | Descriptor: | 1,2-ETHANEDIOL, 7-[(2-chloranyl-1,3-benzothiazol-6-yl)sulfonyl]-~{N}-[(3-chloranyl-4-phenyl-phenyl)methyl]-7-azaspiro[3.5]nonan-2-amine, Casein kinase II subunit alpha, ... | | Authors: | Kraemer, A, Greco, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-03-20 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of human Casein Kinase II subunit alpha (CK2a1) in complex with allosteric ligand FGC3331
To Be Published
|
|
9EPY
 
 | | Crystal structure of human Casein Kinase II subunit alpha (CK2a1) in complex with allosteric ligand FGC3330 | | Descriptor: | 1,2-ETHANEDIOL, Casein kinase II subunit alpha, SULFATE ION, ... | | Authors: | Kraemer, A, Greco, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-03-20 | | Release date: | 2024-05-01 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Synthesis and evaluation of chemical linchpins for highly selective CK2 alpha targeting.
Eur.J.Med.Chem., 276, 2024
|
|
9EPZ
 
 | | Crystal structure of human Casein Kinase II subunit alpha (CK2a1) in complex with allosteric ligand FGC3337 | | Descriptor: | 1,2-ETHANEDIOL, Casein kinase II subunit alpha, SULFATE ION, ... | | Authors: | Kraemer, A, Greco, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-03-20 | | Release date: | 2024-05-01 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Synthesis and evaluation of chemical linchpins for highly selective CK2 alpha targeting.
Eur.J.Med.Chem., 276, 2024
|
|
9EQ0
 
 | | Crystal structure of human Casein Kinase II subunit alpha (CK2a1) in complex with allosteric ligand FGJG12 | | Descriptor: | Casein kinase II subunit alpha, SULFATE ION, ~{N}-[4-[(3-chloranyl-4-phenyl-phenyl)methylamino]butyl]isoquinoline-5-sulfonamide | | Authors: | Kraemer, A, Greco, F, Gerninghaus, J, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-03-20 | | Release date: | 2024-05-01 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Synthesis and evaluation of chemical linchpins for highly selective CK2 alpha targeting.
Eur.J.Med.Chem., 276, 2024
|
|
9EQ1
 
 | | Crystal structure of human Casein Kinase II subunit alpha (CK2a1) in complex with allosteric ligand FGJM24 | | Descriptor: | 1,2-ETHANEDIOL, Casein kinase II subunit alpha, SULFATE ION, ... | | Authors: | Kraemer, A, Greco, F, Moeckel, J, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-03-20 | | Release date: | 2024-05-01 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Synthesis and evaluation of chemical linchpins for highly selective CK2 alpha targeting.
Eur.J.Med.Chem., 276, 2024
|
|
9EQ3
 
 | | Structure of IgE HMM5 bound to FceRIa cryo-EM class 8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, High affinity immunoglobulin epsilon receptor subunit alpha, ... | | Authors: | Andersen, G.R, Jensen, R.K. | | Deposit date: | 2024-03-20 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Structure of IgE HMM5 bound to FceRIa cryo-EM class 8
To be published
|
|
9EQ4
 
 | | Structure of IgE HMM5 bound to FceRIa cryo-EM class 5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, High affinity immunoglobulin epsilon receptor subunit alpha, ... | | Authors: | Andersen, G.R, Jensen, R.K. | | Deposit date: | 2024-03-20 | | Release date: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (8.4 Å) | | Cite: | Structure of IgE HMM5 bound to FceRIa cryo-EM class 5
To be published
|
|
9EQ7
 
 | | Halobacterium salinarum archaellum filament | | Descriptor: | 2-O-sulfo-beta-D-glucopyranuronic acid-(1-4)-3-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-beta-D-glucopyranuronic acid-(1-4)-beta-D-glucopyranose, Archaellin | | Authors: | Grossman-Haham, I, Shahar, A. | | Deposit date: | 2024-03-21 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Perturbed N-glycosylation of Halobacterium salinarum archaellum filaments leads to filament bundling and compromised cell motility.
Nat Commun, 15, 2024
|
|
9EQJ
 
 | | Crystal structure of pVHL:EloB:EloC in complex with MP-1-39 | | Descriptor: | (2S,4R)-1-[(2R)-2-[(1-fluoranylcyclopropyl)carbonylamino]-3-methyl-3-[[cis-4-(morpholin-4-ylmethyl)cyclohexyl]methylsulfanyl]butanoyl]-N-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Kroupova, A, Pierri, M, Liu, X, Ciulli, A. | | Deposit date: | 2024-03-21 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Stereochemical inversion at a 1,4-cyclohexyl PROTAC linker fine-tunes conformation and binding affinity.
Bioorg.Med.Chem.Lett., 110, 2024
|
|
9EQM
 
 | | Crystal structure of pVHL:EloB:EloC in complex with MP-1-21 | | Descriptor: | (2S,4R)-1-[(2R)-2-[(1-fluoranylcyclopropyl)carbonylamino]-3-methyl-3-[[trans-4-(morpholin-4-ylmethyl)cyclohexyl]methylsulfanyl]butanoyl]-N-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Kroupova, A, Pierri, M, Liu, X, Ciulli, A. | | Deposit date: | 2024-03-21 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Stereochemical inversion at a 1,4-cyclohexyl PROTAC linker fine-tunes conformation and binding affinity.
Bioorg.Med.Chem.Lett., 110, 2024
|
|






