7TOS
 
 | | E. coli 70S ribosomes bound with the ALS/FTD-associated dipeptide repeat protein PR20 | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Loveland, A.B, Svidritskiy, E, Susorov, D, Lee, S, Park, A, Zvornicanin, S, Demo, G, Gao, F.B, Korostelev, A.A. | | 登録日 | 2022-01-24 | | 公開日 | 2022-05-25 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Ribosome inhibition by C9ORF72-ALS/FTD-associated poly-PR and poly-GR proteins revealed by cryo-EM.
Nat Commun, 13, 2022
|
|
7TR6
 
 | | Cascade complex from type I-A CRISPR-Cas system | | 分子名称: | Cas11a, Cas5a, Cas7a, ... | | 著者 | Hu, C, Ni, D, Nam, K.H, Majumdar, S, McLean, J, Stahlberg, H, Terns, M, Ke, A. | | 登録日 | 2022-01-28 | | 公開日 | 2022-08-10 | | 最終更新日 | 2022-08-17 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Allosteric control of type I-A CRISPR-Cas3 complexes and establishment as effective nucleic acid detection and human genome editing tools.
Mol.Cell, 82, 2022
|
|
7TR8
 
 | | Cascade complex from type I-A CRISPR-Cas system | | 分子名称: | CRISPR-associated endonuclease Cas3-HD, CRISPR-associated helicase Cas3, Cas11a, ... | | 著者 | Hu, C, Ni, D, Nam, K.H, Majumdar, S, McLean, J, Stahlberg, H, Terns, M, Ke, A. | | 登録日 | 2022-01-28 | | 公開日 | 2022-08-10 | | 最終更新日 | 2022-08-17 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Allosteric control of type I-A CRISPR-Cas3 complexes and establishment as effective nucleic acid detection and human genome editing tools.
Mol.Cell, 82, 2022
|
|
7TR9
 
 | | Cascade complex from type I-A CRISPR-Cas system | | 分子名称: | CRISPR-associated endonuclease Cas3-HD, CRISPR-associated helicase Cas3, Cas11a, ... | | 著者 | Hu, C, Ni, D, Nam, K.H, Majumdar, S, McLean, J, Stahlberg, H, Terns, M, Ke, A. | | 登録日 | 2022-01-28 | | 公開日 | 2022-08-10 | | 最終更新日 | 2022-08-24 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Allosteric control of type I-A CRISPR-Cas3 complexes and establishment as effective nucleic acid detection and human genome editing tools.
Mol.Cell, 82, 2022
|
|
7TRA
 
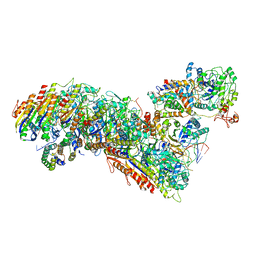 | | Cascade complex from type I-A CRISPR-Cas system | | 分子名称: | CRISPR-associated endonuclease Cas3-HD, CRISPR-associated helicase Cas3, Cas11a, ... | | 著者 | Hu, C, Ni, D, Nam, K.H, Majumdar, S, McLean, J, Stahlberg, H, Terns, M, Ke, A. | | 登録日 | 2022-01-28 | | 公開日 | 2022-08-10 | | 最終更新日 | 2022-08-17 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Allosteric control of type I-A CRISPR-Cas3 complexes and establishment as effective nucleic acid detection and human genome editing tools.
Mol.Cell, 82, 2022
|
|
7TUT
 
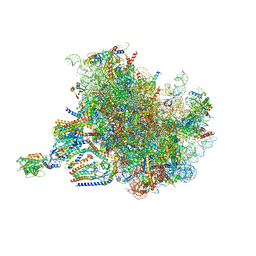 | | Structure of the rabbit 80S ribosome stalled on a 4-TMD Rhodopsin intermediate in complex with the multipass translocon | | 分子名称: | 28S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | 著者 | Kim, M.K, Lewis, A.J.O, Keenan, R.J, Hegde, R.S. | | 登録日 | 2022-02-03 | | 公開日 | 2022-10-19 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.88 Å) | | 主引用文献 | Mechanism of an intramembrane chaperone for multipass membrane proteins.
Nature, 611, 2022
|
|
7TW2
 
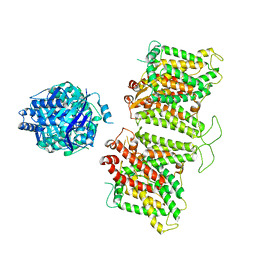 | |
7TZC
 
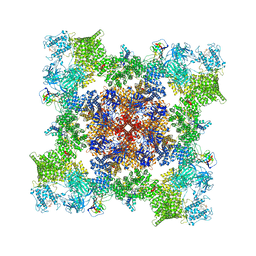 | | A drug and ATP binding site in type 1 ryanodine receptor | | 分子名称: | (2S)-3-(octadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 4-[(7-methoxy-2,3-dihydro-1,4-benzothiazepin-4(5H)-yl)methyl]benzoic acid, ADENOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Melville, Z, Dridi, H, Yuan, Q, Reiken, S, Anetta, W, Liu, Y, Clarke, O.B, Marks, A.R. | | 登録日 | 2022-02-15 | | 公開日 | 2022-05-18 | | 最終更新日 | 2022-07-20 | | 実験手法 | ELECTRON MICROSCOPY (2.45 Å) | | 主引用文献 | A drug and ATP binding site in type 1 ryanodine receptor.
Structure, 30, 2022
|
|
7U05
 
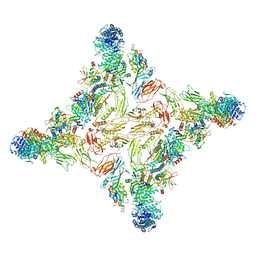 | |
7U06
 
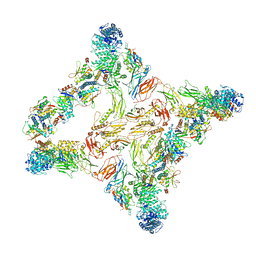 | |
7U0H
 
 | | State NE1 nucleolar 60S ribosome biogenesis intermediate - Overall model | | 分子名称: | 25S rRNA, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, 5.8S rRNA, ... | | 著者 | Cruz, V.E, Sekulski, K, Peddada, N, Erzberger, J.P. | | 登録日 | 2022-02-18 | | 公開日 | 2022-12-14 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (2.76 Å) | | 主引用文献 | Sequence-specific remodeling of a topologically complex RNP substrate by Spb4.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7U0L
 
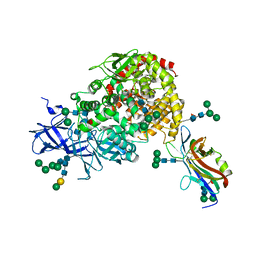 | | Crystal structure of the CCoV-HuPn-2018 RBD (domain B) in complex with canine APN | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Aminopeptidase N, ... | | 著者 | Tortorici, M.A, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2022-02-18 | | 公開日 | 2022-08-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Structure, receptor recognition, and antigenicity of the human coronavirus CCoV-HuPn-2018 spike glycoprotein.
Cell, 185, 2022
|
|
7U0P
 
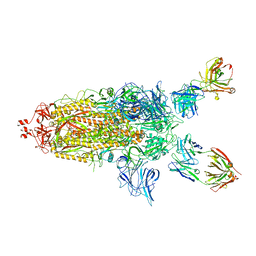 | | SARS-Cov2 S protein structure in complex with neutralizing monoclonal antibody 002-S21F2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Patel, A, Ortlund, E. | | 登録日 | 2022-02-18 | | 公開日 | 2022-08-10 | | 最終更新日 | 2023-02-22 | | 実験手法 | ELECTRON MICROSCOPY (3.76 Å) | | 主引用文献 | Structural insights for neutralization of Omicron variants BA.1, BA.2, BA.4, and BA.5 by a broadly neutralizing SARS-CoV-2 antibody.
Sci Adv, 8, 2022
|
|
7U0Q
 
 | |
7U1I
 
 | |
7U1J
 
 | |
7U2H
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with mRNA, aminoacylated A-site Gly-NH-tRNAgly, aminoacylated P-site fMet-NH-tRNAmet, and deacylated E-site tRNAgly at 2.55A resolution | | 分子名称: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Syroegin, E.A, Aleksandrova, E.V, Polikanov, Y.S. | | 登録日 | 2022-02-24 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Structural basis for the inability of chloramphenicol to inhibit peptide bond formation in the presence of A-site glycine.
Nucleic Acids Res., 50, 2022
|
|
7U2I
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with mRNA, aminoacylated A-site Gly-NH-tRNAgly, aminoacylated P-site fMet-NH-tRNAmet, deacylated E-site tRNAgly, and chloramphenicol at 2.55A resolution | | 分子名称: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Syroegin, E.A, Aleksandrova, E.V, Polikanov, Y.S. | | 登録日 | 2022-02-24 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Structural basis for the inability of chloramphenicol to inhibit peptide bond formation in the presence of A-site glycine.
Nucleic Acids Res., 50, 2022
|
|
7U2J
 
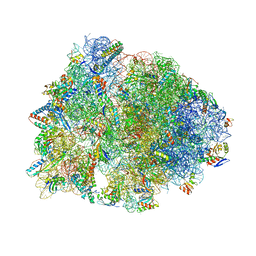 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with mRNA, aminoacylated A-site Gly-NH-tRNAgly, peptidyl P-site fMAC-NH-tRNAmet, deacylated E-site tRNAgly, and chloramphenicol at 2.55A resolution | | 分子名称: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Syroegin, E.A, Aleksandrova, E.V, Polikanov, Y.S. | | 登録日 | 2022-02-24 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Structural basis for the inability of chloramphenicol to inhibit peptide bond formation in the presence of A-site glycine.
Nucleic Acids Res., 50, 2022
|
|
7U3Q
 
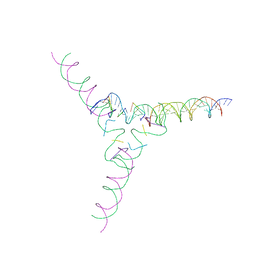 | | [4T7] Self-assembling tensegrity triangle with four turns of DNA per axis with R3 symmetry | | 分子名称: | DNA (29-MER), DNA (42-MER), DNA (5'-D(P*AP*CP*A)-3'), ... | | 著者 | Woloszyn, K, Vecchioni, S, Lu, B, Ma, Y, Seeman, N.C, Sha, R, Ohayon, Y.P, Huang, Q. | | 登録日 | 2022-02-28 | | 公開日 | 2022-09-28 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (9.32 Å) | | 主引用文献 | Augmented DNA Nanoarchitectures: A Structural Library of 3D Self-Assembling Tensegrity Triangle Variants.
Adv Mater, 34, 2022
|
|
7U3V
 
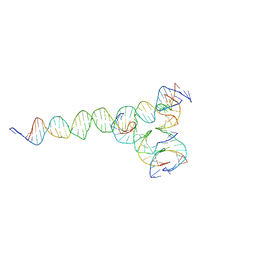 | | [F224] Self-assembling tensegrity triangle with two turns, two turns and four turns of DNA per axis by extension with P1 symmetry | | 分子名称: | DNA (35-MER), DNA (42-MER), DNA (5'-D(*CP*AP*CP*GP*AP*GP*CP*CP*TP*GP*AP*TP*CP*GP*GP*AP*CP*AP*AP*GP*A)-3'), ... | | 著者 | Woloszyn, K, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | 登録日 | 2022-02-28 | | 公開日 | 2022-09-28 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (5.14 Å) | | 主引用文献 | Augmented DNA Nanoarchitectures: A Structural Library of 3D Self-Assembling Tensegrity Triangle Variants.
Adv Mater, 34, 2022
|
|
7U3W
 
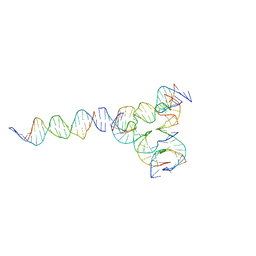 | | [L224] Self-assembling tensegrity triangle with two turns, two turns and four turns of DNA per axis by linker addition with P1 symmetry | | 分子名称: | DNA (5'-D(*AP*AP*CP*CP*TP*AP*CP*CP*TP*GP*GP*CP*AP*GP*GP*AP*CP*GP*AP*CP*T)-3'), DNA (5'-D(*AP*GP*AP*GP*TP*CP*GP*TP*GP*GP*CP*TP*CP*G)-3'), DNA (5'-D(*CP*AP*CP*GP*AP*GP*CP*CP*TP*GP*AP*TP*CP*GP*GP*AP*CP*AP*AP*GP*A)-3'), ... | | 著者 | Woloszyn, K, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | 登録日 | 2022-02-28 | | 公開日 | 2022-09-28 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (6.33 Å) | | 主引用文献 | Augmented DNA Nanoarchitectures: A Structural Library of 3D Self-Assembling Tensegrity Triangle Variants.
Adv Mater, 34, 2022
|
|
7U3Z
 
 | | [F244] Self-assembling tensegrity triangle with two turns, four turns and four turns of DNA per axis by extension with P1 symmetry | | 分子名称: | DNA (35-MER), DNA (42-MER), DNA (5'-D(*AP*AP*CP*CP*TP*AP*CP*CP*TP*GP*GP*CP*AP*GP*GP*AP*CP*GP*AP*CP*T)-3'), ... | | 著者 | Woloszyn, K, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | 登録日 | 2022-02-28 | | 公開日 | 2022-09-28 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (7.55 Å) | | 主引用文献 | Augmented DNA Nanoarchitectures: A Structural Library of 3D Self-Assembling Tensegrity Triangle Variants.
Adv Mater, 34, 2022
|
|
7U40
 
 | | [L244] Self-assembling tensegrity triangle with two turns, four turns and four turns of DNA per axis by linker addition with P1 symmetry | | 分子名称: | DNA (5'-D(*AP*AP*CP*CP*TP*AP*CP*CP*TP*GP*GP*CP*AP*GP*GP*AP*CP*GP*AP*CP*T)-3'), DNA (5'-D(*TP*TP*AP*GP*TP*CP*GP*TP*GP*GP*CP*TP*CP*G)-3'), DNA (5'-D(P*AP*CP*TP*GP*AP*TP*GP*TP*GP*GP*TP*AP*GP*G)-3'), ... | | 著者 | Woloszyn, K, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | 登録日 | 2022-02-28 | | 公開日 | 2022-09-28 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (7.55 Å) | | 主引用文献 | Augmented DNA Nanoarchitectures: A Structural Library of 3D Self-Assembling Tensegrity Triangle Variants.
Adv Mater, 34, 2022
|
|
7U41
 
 | | [F234] Self-assembling tensegrity triangle with two turns, three turns and four turns of DNA per axis by extension with P1 symmetry | | 分子名称: | DNA (31-MER), DNA (35-MER), DNA (42-MER), ... | | 著者 | Woloszyn, K, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | 登録日 | 2022-02-28 | | 公開日 | 2022-09-28 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (7.24 Å) | | 主引用文献 | Augmented DNA Nanoarchitectures: A Structural Library of 3D Self-Assembling Tensegrity Triangle Variants.
Adv Mater, 34, 2022
|
|






