6YJT
 
 | | Crystal structure of MGAT5 (alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase V) luminal domain with a Lys329-Ile345 loop truncation, in complex with UDP | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase A, ... | | Authors: | Wu, L, Darby, J.F, Gilio, A.K, Davies, G.J. | | Deposit date: | 2020-04-04 | | Release date: | 2020-08-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Substrate Engagement and Catalytic Mechanisms of N-Acetylglucosaminyltransferase V
Acs Catalysis, 2020
|
|
6YJU
 
 | | Crystal structure of MGAT5 (alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase V) luminal domain with a Lys329-Ile345 loop truncation, in complex with UDP and biantennary pentasaccharide M592 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose, ... | | Authors: | Wu, L, Darby, J.F, Gilio, A.K, Davies, G.J. | | Deposit date: | 2020-04-04 | | Release date: | 2020-08-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Substrate Engagement and Catalytic Mechanisms of N-Acetylglucosaminyltransferase V
Acs Catalysis, 2020
|
|
6YJV
 
 | | Crystal structure of unliganded MGAT5 (alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase V) luminal domain with a Lys329-Ile345 loop truncation, in complex with UDP-2-deoxy-2-fluoroglucose and biantennary pentasaccharide M592 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose, ... | | Authors: | Wu, L, Darby, J.F, Gilio, A.K, Davies, G.J. | | Deposit date: | 2020-04-04 | | Release date: | 2020-08-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Substrate Engagement and Catalytic Mechanisms of N-Acetylglucosaminyltransferase V
Acs Catalysis, 2020
|
|
6YKG
 
 | |
6YL3
 
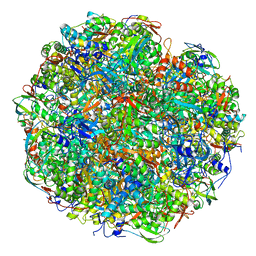 | | High resolution cryo-EM structure of urease from the pathogen Yersinia enterocolitica | | Descriptor: | NICKEL (II) ION, Urease subunit alpha, Urease subunit beta, ... | | Authors: | Righetto, R.D, Anton, L, Adaixo, R, Jakob, R, Zivanov, J, Mahi, M.A, Ringler, P, Schwede, T, Maier, T, Stahlberg, H. | | Deposit date: | 2020-04-06 | | Release date: | 2020-05-06 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (1.98 Å) | | Cite: | High-resolution cryo-EM structure of urease from the pathogen Yersinia enterocolitica.
Nat Commun, 11, 2020
|
|
6YLF
 
 | |
6YLG
 
 | |
6YLH
 
 | |
6YLX
 
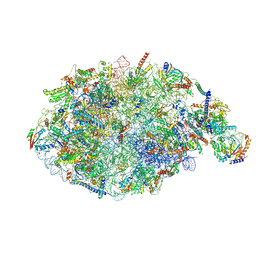 | | pre-60S State NE1 (TAP-Flag-Nop53) | | Descriptor: | 25S rRNA, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, 5.8S rRNA, ... | | Authors: | Kater, L, Beckmann, R. | | Deposit date: | 2020-04-07 | | Release date: | 2020-07-29 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Construction of the Central Protuberance and L1 Stalk during 60S Subunit Biogenesis.
Mol.Cell, 79, 2020
|
|
6YLY
 
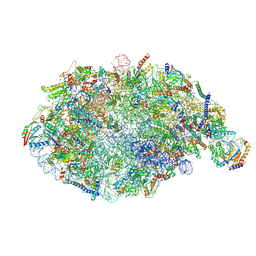 | | pre-60S State NE2 (TAP-Flag-Nop53) | | Descriptor: | 25S rRNA, 5.8S rRNA, 60S ribosomal protein L13-A, ... | | Authors: | Kater, L, Beckmann, R. | | Deposit date: | 2020-04-07 | | Release date: | 2020-07-29 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Construction of the Central Protuberance and L1 Stalk during 60S Subunit Biogenesis.
Mol.Cell, 79, 2020
|
|
6YLZ
 
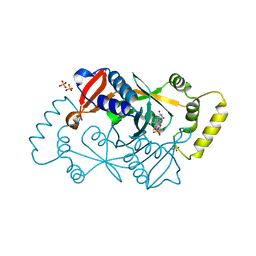 | | X-ray structure of the K72I,Y129F,R133L, H199A quadruple mutant of PNP-oxidase from E. coli | | Descriptor: | FLAVIN MONONUCLEOTIDE, PHOSPHATE ION, Pyridoxine/pyridoxamine 5'-phosphate oxidase, ... | | Authors: | Battista, T, Sularea, M, Barile, A, Fiorillo, A, Tramonti, A, Ilari, A. | | Deposit date: | 2020-04-07 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.558 Å) | | Cite: | Identification and characterization of the pyridoxal 5'-phosphate allosteric site in Escherichia coli pyridoxine 5'-phosphate oxidase.
J.Biol.Chem., 296, 2021
|
|
6YMH
 
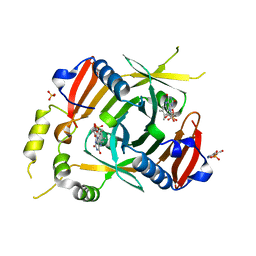 | | X-ray structure of the K72I, Y129F, R133L, H199A quadruple mutant of PNP-oxidase from E. coli in complex with PLP | | Descriptor: | FLAVIN MONONUCLEOTIDE, PYRIDOXAL-5'-PHOSPHATE, Pyridoxine/pyridoxamine 5'-phosphate oxidase, ... | | Authors: | Battista, T, Sularea, M, Barile, A, Fiorillo, A, Tramonti, A, Ilari, A. | | Deposit date: | 2020-04-08 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.417 Å) | | Cite: | Identification and characterization of the pyridoxal 5'-phosphate allosteric site in Escherichia coli pyridoxine 5'-phosphate oxidase.
J.Biol.Chem., 296, 2021
|
|
6YMN
 
 | |
6YMQ
 
 | | TREM2 extracellular domain (19-131) in complex with single-chain variable 4 (scFv-4) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Single-chain variable 4, ... | | Authors: | Szykowska, A, Preger, C, Scacioc, A, Mukhopadhyay, S.M.M, McKinley, G, Graslund, S, Wigren, E, Persson, H, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Di Daniel, E, Davis, J.B, Burgess-Brown, N, Bullock, A. | | Deposit date: | 2020-04-09 | | Release date: | 2021-02-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Selection and structural characterization of anti-TREM2 scFvs that reduce levels of shed ectodomain.
Structure, 29, 2021
|
|
6YN6
 
 | | Inducible lysine decarboxylase LdcI stacks, pH 5.7 | | Descriptor: | Inducible lysine decarboxylase | | Authors: | Felix, J, Jessop, M, Desfosses, A, Effantin, G, Gutsche, I. | | Deposit date: | 2020-04-10 | | Release date: | 2021-01-13 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Supramolecular assembly of the Escherichia coli LdcI upon acid stress.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6YNX
 
 | | Cryo-EM structure of Tetrahymena thermophila mitochondrial ATP synthase - Fo-subcomplex | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Kock Flygaard, R, Muhleip, A, Amunts, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-09-30 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Type III ATP synthase is a symmetry-deviated dimer that induces membrane curvature through tetramerization.
Nat Commun, 11, 2020
|
|
6YNY
 
 | | Cryo-EM structure of Tetrahymena thermophila mitochondrial ATP synthase - F1Fo composite dimer model | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Kock Flygaard, R, Muhleip, A, Amunts, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-09-30 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Type III ATP synthase is a symmetry-deviated dimer that induces membrane curvature through tetramerization.
Nat Commun, 11, 2020
|
|
6YNZ
 
 | | Cryo-EM structure of Tetrahymena thermophila mitochondrial ATP synthase - F1Fo composite tetramer model | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Kock Flygaard, R, Muhleip, A, Amunts, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-09-30 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Type III ATP synthase is a symmetry-deviated dimer that induces membrane curvature through tetramerization.
Nat Commun, 11, 2020
|
|
6YO0
 
 | | Cryo-EM structure of Tetrahymena thermophila mitochondrial ATP synthase - F1/peripheral stalk | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase subunit alpha, ... | | Authors: | Kock Flygaard, R, Muhleip, A, Amunts, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-09-30 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Type III ATP synthase is a symmetry-deviated dimer that induces membrane curvature through tetramerization.
Nat Commun, 11, 2020
|
|
6YO5
 
 | | Crystal structure of the M295F variant of Ssl1 | | Descriptor: | ALA-HIS-ALA, COPPER (II) ION, Copper oxidase, ... | | Authors: | Mielenbrink, S, Olbrich, A, Urlacher, V, Span, I. | | Deposit date: | 2020-04-14 | | Release date: | 2021-04-28 | | Last modified: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Substitution of the axial Type 1 Cu Ligand Afford Binding of a Water Molecule in Axial Position Affecting Kinetics, Spectral, and Structural Properties of the Small Laccase Ssl1.
Chemistry, 2024
|
|
6YOZ
 
 | | HiCel7B labelled with b-1,4-glucosyl cyclophellitol | | Descriptor: | (1R,2S,3S,4S,5R,6R)-6-(HYDROXYMETHYL)CYCLOHEXANE-1,2,3,4,5-PENTOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETAMIDE, ... | | Authors: | McGregor, N.G.S, Davies, G.J. | | Deposit date: | 2020-04-15 | | Release date: | 2020-09-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Glycosylated cyclophellitol-derived activity-based probes and inhibitors for cellulases.
Rsc Chem Biol, 1, 2020
|
|
6YP1
 
 | | HiCel7B unliganded | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETAMIDE, Endoglucanase 1, ... | | Authors: | McGregor, N.G.S, Davies, G.J. | | Deposit date: | 2020-04-15 | | Release date: | 2020-09-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Glycosylated cyclophellitol-derived activity-based probes and inhibitors for cellulases.
Rsc Chem Biol, 1, 2020
|
|
6YPR
 
 | | Human histidine triad nucleotide-binding protein 2 (hHINT2) refined to 1.26 A in H32 space group | | Descriptor: | GLYCEROL, Histidine triad nucleotide-binding protein 2, mitochondrial | | Authors: | Dolot, R.D, Wlodarczyk, A, Bujacz, G.D, Nawrot, B.C. | | Deposit date: | 2020-04-16 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Biochemical, crystallographic and biophysical characterization of histidine triad nucleotide-binding protein 2 with different ligands including a non-hydrolyzable analog of Ap4A.
Biochim Biophys Acta Gen Subj, 1865, 2021
|
|
6YPX
 
 | | Human histidine triad nucleotide-binding protein 2 (hHINT2) refined to 2.11 A in C2221 space group | | Descriptor: | CACODYLATE ION, GLYCEROL, Histidine triad nucleotide-binding protein 2, ... | | Authors: | Dolot, R.D, Wlodarczyk, A, Bujacz, G.D, Nawrot, B.C. | | Deposit date: | 2020-04-16 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Biochemical, crystallographic and biophysical characterization of histidine triad nucleotide-binding protein 2 with different ligands including a non-hydrolyzable analog of Ap4A.
Biochim Biophys Acta Gen Subj, 1865, 2021
|
|
6YPZ
 
 | | Promiscuous Reductase LugOII Catalyzes Keto-reduction at C1 during Lugdunomycin Biosynthesis | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Monooxygenase, ... | | Authors: | Xiao, X, Elsayed, S.S, Wu, C, van der Heul, H, Prota, A, Huang, J, Guo, R, Abrahams, J.P, van Wezel, G.P. | | Deposit date: | 2020-04-16 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Functional and Structural Insights into a Novel Promiscuous Ketoreductase of the Lugdunomycin Biosynthetic Pathway.
Acs Chem.Biol., 15, 2020
|
|







