8HPQ
 
 | |
8HPV
 
 | |
8HQF
 
 | | Crystal structure of SARS-Cov-2 main protease G15S mutant in complex with inhibitor YH-53 | | Descriptor: | N-[(2S)-1-[[(2S)-1-(1,3-benzothiazol-2-yl)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]-4-methoxy-1H-indole-2-carboxamide, Non-structural protein 7 | | Authors: | Zeng, P, Zhang, J, Li, J. | | Deposit date: | 2022-12-13 | | Release date: | 2024-01-31 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Crystal structures of main proteases of SARS-CoV-2 variants bound to a benzothiazole-based inhibitor.
Acta Biochim.Biophys.Sin., 55, 2023
|
|
8I4H
 
 | | Omicron spike variant BA.1 with Bn03 | | Descriptor: | Bn03, Spike glycoprotein | | Authors: | Hao, A.H, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2023-01-19 | | Release date: | 2024-01-31 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Defining a highly conserved cryptic epitope for antibody recognition of SARS-CoV-2 variants.
Signal Transduct Target Ther, 8, 2023
|
|
8KA8
 
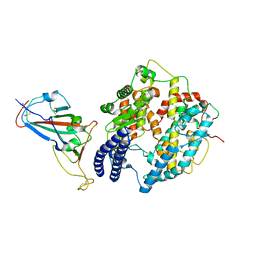 | | Cryo-EM structure of SARS-CoV-2 Delta RBD in complex with golden hamster ACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Niu, S, Zhao, Z.N, Chai, Y, Gao, G.F. | | Deposit date: | 2023-08-02 | | Release date: | 2024-01-31 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Structural basis and analysis of hamster ACE2 binding to different SARS-CoV-2 spike RBDs.
J.Virol., 98, 2024
|
|
8KC2
 
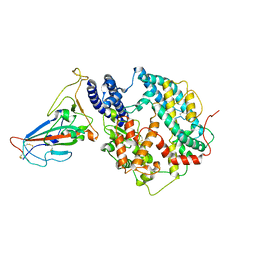 | | Cryo-EM structure of SARS-CoV-2 BA.3 RBD in complex with golden hamster ACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Niu, S, Zhao, Z.N, Chai, Y, Gao, G.F. | | Deposit date: | 2023-08-05 | | Release date: | 2024-01-31 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis and analysis of hamster ACE2 binding to different SARS-CoV-2 spike RBDs.
J.Virol., 98, 2024
|
|
8PH4
 
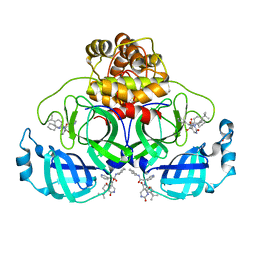 | | Co-Crystal structure of the SARS-CoV2 main protease Nsp5 with an Uracil-carrying X77-like inhibitor | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, MALONATE ION, ... | | Authors: | Barthel, T, Altincekic, N, Jores, N, Wollenhaupt, J, Weiss, M.S, Schwalbe, H. | | Deposit date: | 2023-06-18 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Targeting the Main Protease (M pro , nsp5) by Growth of Fragment Scaffolds Exploiting Structure-Based Methodologies.
Acs Chem.Biol., 19, 2024
|
|
8QPR
 
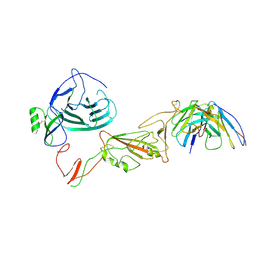 | | SARS-CoV-2 S protein bound to human neutralising antibody UZGENT_G5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, IgG heavy chain - FAB, ... | | Authors: | Remaut, H, Reiter, D, Vandenkerckhove, L, Acar, D.D, Witkowski, W, Gerlo, S. | | Deposit date: | 2023-10-03 | | Release date: | 2024-01-31 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Integrating artificial intelligence-based epitope prediction in a SARS-CoV-2 antibody discovery pipeline: caution is warranted.
Ebiomedicine, 100, 2024
|
|
8QQ0
 
 | | SARS-CoV-2 S protein bound to neutralising antibody UZGENT_A3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Remaut, H, Reiter, D, Vandenkerckhove, L, Acar, D.D, Witkowski, W, Gerlo, S. | | Deposit date: | 2023-10-03 | | Release date: | 2024-01-31 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Integrating artificial intelligence-based epitope prediction in a SARS-CoV-2 antibody discovery pipeline: caution is warranted.
Ebiomedicine, 100, 2024
|
|
8C9L
 
 | |
8C9O
 
 | | Crystal structure of SARS-CoV-2 Mpro-S144A mutant in complex with 13b-K | | Descriptor: | 3C-like proteinase nsp5, ~{tert}-butyl ~{N}-[1-[(2~{S})-3-cyclopropyl-1-oxidanylidene-1-[[(2~{S},3~{R})-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | Authors: | Ibrahim, M, El Kilani, H, Hilgenfeld, R. | | Deposit date: | 2023-01-23 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal structure of SARS-CoV-2 Mpro_S144A mutant in complex with 13b-K
To Be Published
|
|
8CBD
 
 | | SARS-CoV-2 Delta-RBD complexed with BA.4/5-1 and EY6A Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BA.4/5-1 heavy chain, BA.4/5-1 light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2023-01-25 | | Release date: | 2024-02-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.52 Å) | | Cite: | Emerging variants develop total escape from potent monoclonal antibodies induced by BA.4/5 infection.
Nat Commun, 15, 2024
|
|
8CBE
 
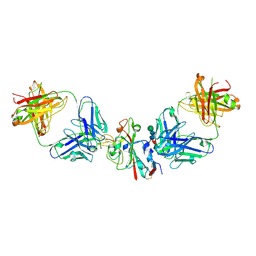 | | SARS-CoV-2 Delta-RBD complexed with BA.4/5-2 and Beta-49 Fabs | | Descriptor: | BA.4/5-2 heavy chain, BA.4/5-2 light chain, Beta-49 heavy chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2023-01-25 | | Release date: | 2024-02-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Emerging variants develop total escape from potent monoclonal antibodies induced by BA.4/5 infection.
Nat Commun, 15, 2024
|
|
8CBF
 
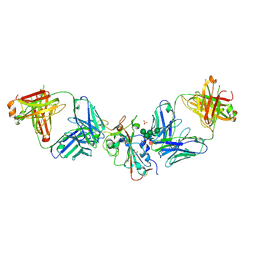 | | SARS-CoV-2 Delta-RBD complexed with Omi-42 and Beta-49 Fabs | | Descriptor: | Beta-49 heavy chain, Beta-49 light chain, CHLORIDE ION, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2023-01-25 | | Release date: | 2024-02-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Emerging variants develop total escape from potent monoclonal antibodies induced by BA.4/5 infection.
Nat Commun, 15, 2024
|
|
8DT9
 
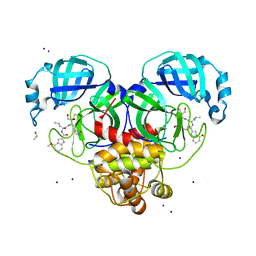 | | Crystal Structure of SARS CoV-2 Mpro mutant L141R with Pfizer Intravenous Inhibitor PF-00835231 | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide, ... | | Authors: | Shaqra, A.M, Schiffer, C.A. | | Deposit date: | 2022-07-25 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Contributions of Hyperactive Mutations in M pro from SARS-CoV-2 to Drug Resistance.
Acs Infect Dis., 10, 2024
|
|
8E4W
 
 | |
8E5C
 
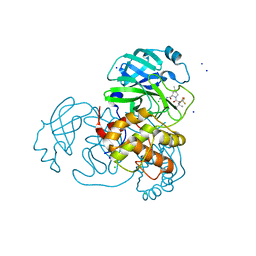 | | Crystal Structure of SARS CoV-2 Mpro mutant L50F with Nirmatrelvir captured in two conformational states | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Shaqra, A.M, Schiffer, C.A. | | Deposit date: | 2022-08-20 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Contributions of Hyperactive Mutations in M pro from SARS-CoV-2 to Drug Resistance.
Acs Infect Dis., 10, 2024
|
|
8I9B
 
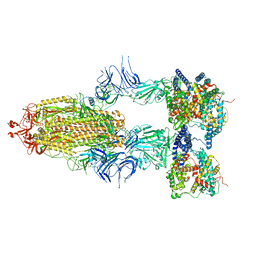 | | S-ECD (Omicron BA.2.75) in complex with PD of ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Li, Y.N, Shen, Y.P, Zhang, Y.Y, Yan, R.H. | | Deposit date: | 2023-02-06 | | Release date: | 2024-02-07 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures of SARS-CoV-2 BA.2-derived subvariants spike in complex with ACE2 receptor.
Cell Discov, 9, 2023
|
|
8I9C
 
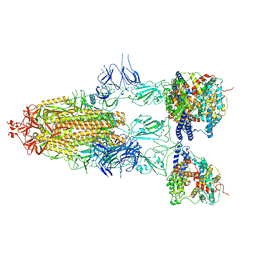 | | S-ECD (Omicron BF.7) in complex with PD of ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Li, Y.N, Shen, Y.P, Zhang, Y.Y, Yan, R.H. | | Deposit date: | 2023-02-06 | | Release date: | 2024-02-07 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Cryo-EM structures of SARS-CoV-2 BA.2-derived subvariants spike in complex with ACE2 receptor.
Cell Discov, 9, 2023
|
|
8I9D
 
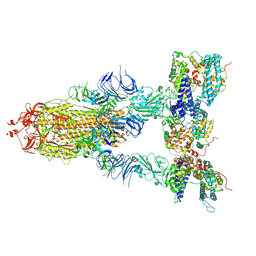 | | S-ECD (Omicron XBB.1) in complex with PD of ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Li, Y.N, Shen, Y.P, Zhang, Y.Y, Yan, R.H. | | Deposit date: | 2023-02-06 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Cryo-EM structures of SARS-CoV-2 BA.2-derived subvariants spike in complex with ACE2 receptor.
Cell Discov, 9, 2023
|
|
8I9E
 
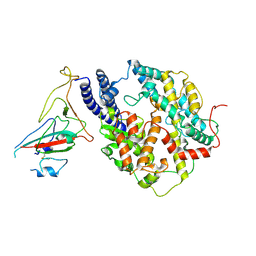 | | S-RBD(Omicron BA.3) in complex with PD of ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Li, Y.N, Shen, Y.P, Zhang, Y.Y, Yan, R.H. | | Deposit date: | 2023-02-06 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural Basis for the Enhanced Infectivity and Immune Evasion of Omicron Subvariants.
Viruses, 15, 2023
|
|
8I9F
 
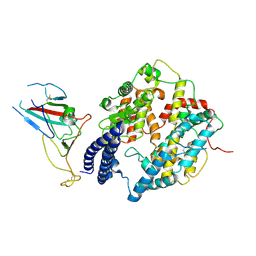 | | S-RBD (Omicron BA.2.75) in complex with PD of ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Li, Y.N, Shen, Y.P, Zhang, Y.Y, Yan, R.H. | | Deposit date: | 2023-02-06 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures of SARS-CoV-2 BA.2-derived subvariants spike in complex with ACE2 receptor.
Cell Discov, 9, 2023
|
|
8I9G
 
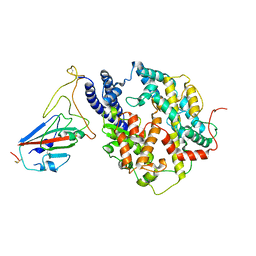 | | S-RBD (Omicron BF.7) in complex with PD of ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Li, Y.N, Shen, Y.P, Zhang, Y.Y, Yan, R.H. | | Deposit date: | 2023-02-06 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of SARS-CoV-2 BA.2-derived subvariants spike in complex with ACE2 receptor.
Cell Discov, 9, 2023
|
|
8I9H
 
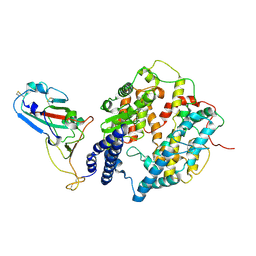 | | S-RBD (Omicron XBB.1) in complex with PD of ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Li, Y.N, Shen, Y.P, Zhang, Y.Y, Yan, R.H. | | Deposit date: | 2023-02-06 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures of SARS-CoV-2 BA.2-derived subvariants spike in complex with ACE2 receptor.
Cell Discov, 9, 2023
|
|
8IQJ
 
 | | Crystal structure of SARS-CoV2 N-NTD | | Descriptor: | Nucleoprotein | | Authors: | Hong, J.Y, Hou, M.H. | | Deposit date: | 2023-03-16 | | Release date: | 2024-02-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Targeting protein-protein interaction interfaces with antiviral N protein inhibitor in SARS-CoV-2.
Biophys.J., 123, 2024
|
|








