8IFY
 
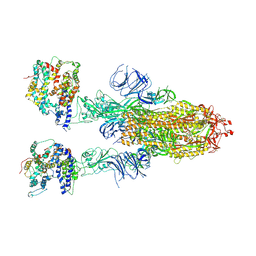 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.4/5 spike protein in complex with white-tailed deer ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Han, P, Meng, Y.M, Qi, J.X. | | Deposit date: | 2023-02-20 | | Release date: | 2023-08-30 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Structural basis of white-tailed deer, Odocoileus virginianus , ACE2 recognizing all the SARS-CoV-2 variants of concern with high affinity.
J.Virol., 97, 2023
|
|
8IFZ
 
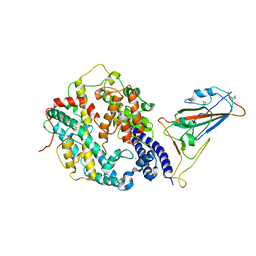 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.4/5 spike protein receptor-binding domain in complex with white-tailed deer ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, Spike protein S1, ... | | Authors: | Han, P, Meng, Y.M, Qi, J.X. | | Deposit date: | 2023-02-20 | | Release date: | 2023-08-30 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural basis of white-tailed deer, Odocoileus virginianus , ACE2 recognizing all the SARS-CoV-2 variants of concern with high affinity.
J.Virol., 97, 2023
|
|
8STY
 
 | |
8STZ
 
 | |
8SXR
 
 | | Crystal structure of SARS-CoV-2 Mpro with C5a | | Descriptor: | 3C-like proteinase nsp5, N-[(4-chlorothiophen-2-yl)methyl]-N-[4-(dimethylamino)phenyl]-2-(5-hydroxyisoquinolin-4-yl)acetamide | | Authors: | Worrall, L.J, Kenward, C, Lee, J, Strynadka, N.C.J. | | Deposit date: | 2023-05-23 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.114 Å) | | Cite: | A novel class of broad-spectrum active-site-directed 3C-like protease inhibitors with nanomolar antiviral activity against highly immune-evasive SARS-CoV-2 Omicron subvariants.
Emerg Microbes Infect, 12, 2023
|
|
7U92
 
 | | SARS-CoV-2 Main Protease (Mpro) in Complex with ML1006a | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-(azetidin-1-yl)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase, CHLORIDE ION | | Authors: | Westberg, M, Fernandez, D, Lin, M.Z. | | Deposit date: | 2022-03-09 | | Release date: | 2023-09-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An orally bioavailable SARS-CoV-2 main protease inhibitor exhibits improved affinity and reduced sensitivity to mutations.
Sci Transl Med, 16, 2024
|
|
7Y0O
 
 | |
7YBK
 
 | | SARS-CoV-2 B.1.620 variant spike (open state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-06-29 | | Release date: | 2023-09-06 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of SARS-CoV-2 spike protein alert noteworthy sites for the potential approaching variants.
Virol Sin, 37, 2022
|
|
7YLB
 
 | |
8GSB
 
 | |
7XY3
 
 | |
7XY4
 
 | |
7Y0N
 
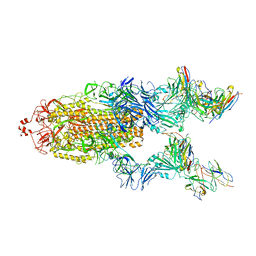 | | SARS-CoV-2 WT Spike in complex with R15 Fab and P14 Nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R15-F7, ... | | Authors: | Wang, L. | | Deposit date: | 2022-06-05 | | Release date: | 2023-09-13 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | SARS-CoV-2 WT Spike in complex with R15 Fab and P14 Nanobody
To Be Published
|
|
8AZC
 
 | |
8AZD
 
 | |
8F0I
 
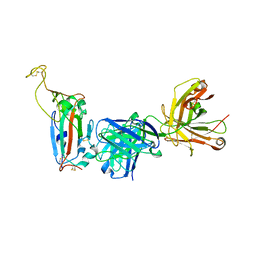 | |
8GTV
 
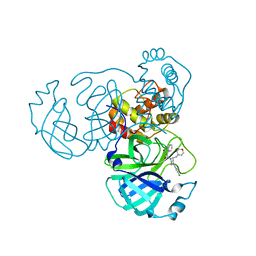 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound JZD-07 | | Descriptor: | 3C-like proteinase, 4-[(2~{S})-4-(3,4-dichlorophenyl)-2-(morpholin-4-ylmethyl)piperazin-1-yl]carbonyl-1~{H}-quinolin-2-one | | Authors: | Su, H.X, Nie, T.Q, Xie, H, Li, M.J, Xu, Y.C. | | Deposit date: | 2022-09-08 | | Release date: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Development of novel inhibitors against SARS-CoV-2 3CLpro
To Be Published
|
|
8GTW
 
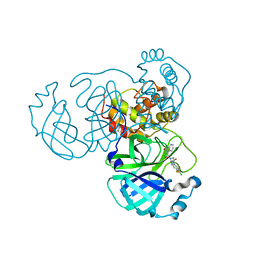 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound JZD-26 | | Descriptor: | (2S)-4-(3,4-dichlorophenyl)-1-[(2-oxidanylidene-1H-quinolin-4-yl)carbonyl]-N-[3,3,3-tris(fluoranyl)propyl]piperazine-2-carboxamide, 3C-like proteinase | | Authors: | Su, H.X, Nie, T.Q, Xie, H, Li, M.J, Xu, Y.C. | | Deposit date: | 2022-09-08 | | Release date: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Development of novel inhibitors against SARS-CoV-2 3CLpro
To Be Published
|
|
8U2X
 
 | |
7YR4
 
 | |
8CQ1
 
 | |
8EDF
 
 | |
8GVD
 
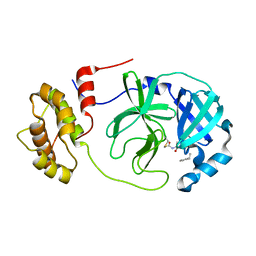 | | SARS-CoV-2 Mpro in complex with D-4-38 | | Descriptor: | 3-PYRIDIN-4-YL-2,4-DIHYDRO-INDENO[1,2-.C.]PYRAZOLE, 3C-like proteinase nsp5 | | Authors: | Liu, M, Huang, H. | | Deposit date: | 2022-09-14 | | Release date: | 2023-09-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | SARS-CoV-2 Mpro in complex with D-4-38
To Be Published
|
|
8GVY
 
 | | SARS CoV-2 Mpro in complex with D-3-149 | | Descriptor: | 3-PYRIDIN-4-YL-2,4-DIHYDRO-INDENO[1,2-.C.]PYRAZOLE, 3C-like proteinase nsp5 | | Authors: | Liu, M, Huang, H. | | Deposit date: | 2022-09-16 | | Release date: | 2023-09-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | SARS CoV-2 Mpro in complex with D-3-149
To Be Published
|
|
8GWJ
 
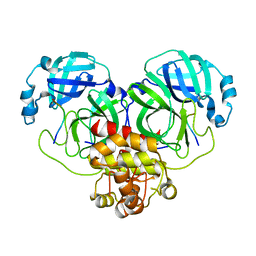 | |








