9CFG
 
 | |
9CFH
 
 | |
9J8T
 
 | |
9J8U
 
 | |
9P6P
 
 | | Crystal Structure of the SARS-CoV-2 2'-O-Methyltransferase with (m7GpppA)pUpU (Cap-0) and S-Adenosyl-L-homocysteine (SAH). | | Descriptor: | 2'-O-methyltransferase, 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Minasov, G, Shuvalova, L, Maltseva, N, Kim, Y, Kiryukhina, O, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2025-06-19 | | Release date: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of the SARS-CoV-2 2'-O-Methyltransferase with (m7GpppA)pUpU (Cap-0) and S-Adenosyl-L-homocysteine (SAH).
To Be Published
|
|
9CJP
 
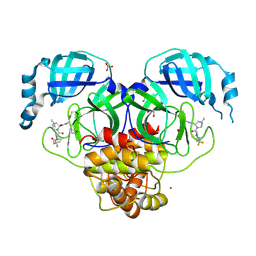 | | X-ray crystal structure of SARS-CoV-2 main protease quadruple mutants in complex with Nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ... | | Authors: | Esler, M.A, Shi, K, Harris, R.S, Aihara, H. | | Deposit date: | 2024-07-07 | | Release date: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | X-ray crystal structure of SARS-CoV-2 main protease quadruple mutants in complex with Nirmatrelvir
To Be Published
|
|
9CJQ
 
 | | X-ray crystal structure of SARS-CoV-2 main protease quadruple mutants in complex with Ensitrelvir | | Descriptor: | 1,2-ETHANEDIOL, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione, main protease | | Authors: | Esler, M.A, Shi, K, Harris, R.S, Aihara, H. | | Deposit date: | 2024-07-07 | | Release date: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | X-ray crystal structure of SARS-CoV-2 main protease quadruple mutants in complex with Ensitrelvir
To Be Published
|
|
9CJR
 
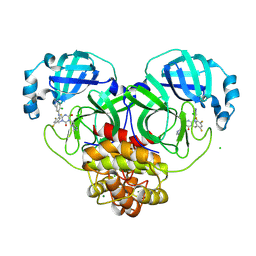 | | X-ray crystal structure of SARS-CoV-2 main protease double mutants in complex with Ensitrelvir | | Descriptor: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione, CHLORIDE ION, ... | | Authors: | Esler, M.A, Shi, K, Harris, R.S, Aihara, H. | | Deposit date: | 2024-07-07 | | Release date: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | X-ray crystal structure of SARS-CoV-2 main protease double mutants in complex with Ensitrelvir
To Be Published
|
|
9CJS
 
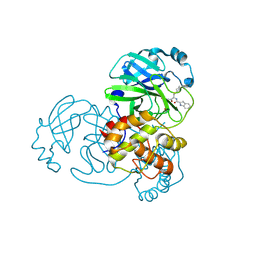 | |
9E9Q
 
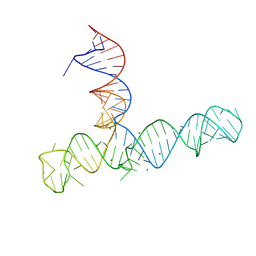 | |
9MPW
 
 | | SARS-CoV2 Spike S2 Subunit in complex with M15 Antibody Fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab M15 Heavy Chain, ... | | Authors: | Sapse, I.A, Bajic, G. | | Deposit date: | 2024-12-31 | | Release date: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | SARS-CoV2 Spike S2 Subunit in complex with M15 Antibody Fragment
To Be Published
|
|
9RCZ
 
 | |
9IQP
 
 | | Crystal structure of the Wuhan SARS-CoV-2 Spike RBD (319-541) complexed with 1p1B10 nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody 1p1B10, SODIUM ION, ... | | Authors: | Sluchanko, N.N, Matyuta, I.O, Dronova, E.A, Favorskaya, I.A, Esmagambetov, I.B, Shcheblyakov, D.V, Logunov, D.Y, Gintsburg, A.L, Popov, V.O, Boyko, K.M. | | Deposit date: | 2024-07-13 | | Release date: | 2025-07-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Ultra-potent RBM-specific single-domain antibody broadly neutralizes multiple SARS-CoV-2 variants with picomolar activity.
Int.J.Biol.Macromol., 319, 2025
|
|
9UXD
 
 | | SARS-CoV2 Spike protein with Fab fragment antibody KXD355,state1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody KXD355, ... | | Authors: | Wang, H. | | Deposit date: | 2025-05-13 | | Release date: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | A rare B cell clonotype imprinted by ancestral SARS-CoV-2 develops cross-sarbecovirus neutralization in immune recalls.
Cell Rep, 44, 2025
|
|
9VCK
 
 | | Cryo-EM structure of SARS-CoV-2 nsp10/nsp14:RNA:SMP complex | | Descriptor: | CALCIUM ION, Guanine-N7 methyltransferase nsp14, Non-structural protein 10, ... | | Authors: | Wang, J, Lou, Z, Liu, D. | | Deposit date: | 2025-06-06 | | Release date: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (4.22 Å) | | Cite: | Structural Basis and Rational Design of Nucleotide Analogue Inhibitor Evading the SARS-CoV-2 Proofreading Enzyme.
J.Am.Chem.Soc., 147, 2025
|
|
9VCL
 
 | | Cryo-EM structure of SARS-CoV-2 nsp10/nsp14:RNA:ATMP complex | | Descriptor: | CALCIUM ION, Guanine-N7 methyltransferase nsp14, Non-structural protein 10, ... | | Authors: | Wang, J, Lou, Z, Liu, D. | | Deposit date: | 2025-06-06 | | Release date: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structural Basis and Rational Design of Nucleotide Analogue Inhibitor Evading the SARS-CoV-2 Proofreading Enzyme.
J.Am.Chem.Soc., 147, 2025
|
|
9CB0
 
 | | SARS-CoV-2 S protein - Accum modified | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Caveney, N.A. | | Deposit date: | 2024-06-18 | | Release date: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | SARS-CoV-2 S protein - Accum modified
To Be Published
|
|
9H6U
 
 | | SARS-CoV-2 S protein in complex with pT1679 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin, pT1679 Fab heavy chain, ... | | Authors: | Hansen, G, Benecke, T, Vollmer, B, Gruenewald, K, Krey, T. | | Deposit date: | 2024-10-25 | | Release date: | 2025-07-23 | | Last modified: | 2025-09-24 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | A critical residue in a conserved RBD epitope determines neutralization breadth of pan-sarbecovirus antibodies with recurring YYDRxxG motifs.
Mbio, 16, 2025
|
|
9MVM
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro)in Complex with Inhibitor AVI-3318 | | Descriptor: | 1-[(4-chlorothiophen-2-yl)methyl]-3-[(2-oxo-1,2-dihydropyridin-3-yl)methyl]-1,3-diazinane-2,4-dione, Replicase polyprotein 1ab | | Authors: | Diallo, A, Gumpena, R, Verba, K. | | Deposit date: | 2025-01-15 | | Release date: | 2025-07-23 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure-based discovery of highly bioavailable, covalent, broad-spectrum coronavirus M Pro inhibitors with potent in vivo efficacy.
Sci Adv, 11, 2025
|
|
9MVQ
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) variant Q192T in Complex with Inhibitor AVI-4303 | | Descriptor: | (3M,5P,6M)-5-(1H-1,2,3-benzotriazol-1-yl)-6-(3-chlorophenyl)-3-(isoquinolin-4-yl)pyrimidine-2,4(1H,3H)-dione, 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ... | | Authors: | Diallo, A, Gumpena, R, Partridge, J, Verba, K. | | Deposit date: | 2025-01-15 | | Release date: | 2025-07-23 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structure-based discovery of highly bioavailable, covalent, broad-spectrum coronavirus M Pro inhibitors with potent in vivo efficacy.
Sci Adv, 11, 2025
|
|
9E21
 
 | |
9I51
 
 | | Crystal structure of the SARS-CoV-2 helicase NSP13 in complex with ADP | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, PHOSPHATE ION, ... | | Authors: | Kloskowski, P, Neumann, P, Ficner, R. | | Deposit date: | 2025-01-27 | | Release date: | 2025-07-30 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Nucleotide-bound crystal structures of the SARS-CoV-2 helicase NSP13.
Acta Crystallogr.,Sect.F, 81, 2025
|
|
9N6N
 
 | |
9VWY
 
 | | Crystal structure of C270S mutant of Papain-like protease (PLpro) from SARS-CoV-2 | | Descriptor: | CHLORIDE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Arya, R, Ganesh, J, Prashar, V, Kumar, M. | | Deposit date: | 2025-07-17 | | Release date: | 2025-07-30 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structure of C270S mutant of Papain-like protease (PLpro) from SARS-CoV-2
To Be Published
|
|
8CAE
 
 | | Crystal structure of SARS-CoV-2 Mpro-H172Y mutant in complex with nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, Non-structural protein 11 | | Authors: | El Kilani, H, Hilgenfeld, R. | | Deposit date: | 2023-01-24 | | Release date: | 2025-08-06 | | Last modified: | 2025-09-03 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | 13b-K and Nirmatrelvir Resistance Mutations of SARS-CoV-2 Main Protease: Structural, Biochemical, and Biophysical Characterization of Free Enzymes and Inhibitor Complexes
Crystals, 15, 2025
|
|








