7WPH
 
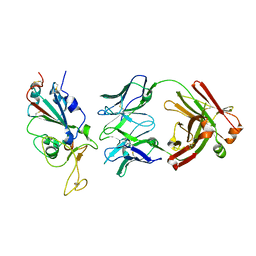 | | SARS-CoV2 RBD bound to Fab06 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FAB06 light chain, Fab06 heavy chain, ... | | Authors: | Lin, J.Q, El Sahili, A, Lescar, J. | | Deposit date: | 2022-01-23 | | Release date: | 2022-03-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Engineering SARS-CoV-2 specific cocktail antibodies into a bispecific format improves neutralizing potency and breadth.
Nat Commun, 13, 2022
|
|
7E9N
 
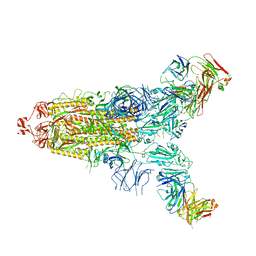 | | Cryo-EM structure of the SARS-CoV-2 S-6P in complex with 35B5 Fab(1 down RBD, state1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 35B5 Fab, Light chain of 35B5 Fab, ... | | Authors: | Wang, X.F, Zhu, Y.Q. | | Deposit date: | 2021-03-04 | | Release date: | 2022-04-06 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | A potent human monoclonal antibody with pan-neutralizing activities directly dislocates S trimer of SARS-CoV-2 through binding both up and down forms of RBD
Signal Transduct Target Ther, 7, 2022
|
|
7ENF
 
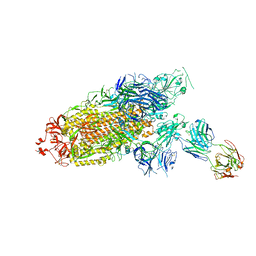 | | Cryo-EM structure of the SARS-CoV-2 S-6P in complex with Fab30 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab30, ... | | Authors: | Wang, X.F, Zhu, Y.Q. | | Deposit date: | 2021-04-16 | | Release date: | 2022-04-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | A potent human monoclonal antibody with pan-neutralizing activities directly dislocates S trimer of SARS-CoV-2 through binding both up and down forms of RBD
Signal Transduct Target Ther, 7, 2022
|
|
7ENG
 
 | |
7RZQ
 
 | |
7RZR
 
 | |
7RZS
 
 | |
7RZT
 
 | |
7RZU
 
 | |
7RZV
 
 | |
7TIK
 
 | |
7VAH
 
 | |
7VLP
 
 | | Crystal structure of SARS-Cov-2 main protease in complex with PF07321332 in spacegroup P1211 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, Replicase polyprotein 1a | | Authors: | Zhou, X.L, Zhong, F.L, Lin, C, Li, J, Zhang, J. | | Deposit date: | 2021-10-05 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.50251932385 Å) | | Cite: | Structural Basis of the Main Proteases of Coronavirus Bound to Drug Candidate PF-07321332.
J.Virol., 96, 2022
|
|
7VLQ
 
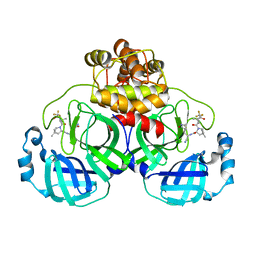 | | Crystal structure of SARS-Cov-2 main protease in complex with PF07321332 in spacegroup P212121 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Zhou, X.L, Zhong, F.L, Lin, C, Zhang, J, Li, J. | | Deposit date: | 2021-10-05 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93910601486 Å) | | Cite: | Structural Basis of the Main Proteases of Coronavirus Bound to Drug Candidate PF-07321332.
J.Virol., 96, 2022
|
|
7VTH
 
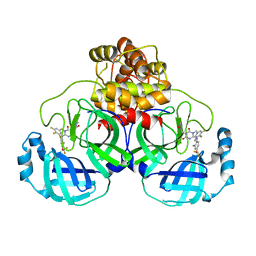 | | The crystal structure of SARS-CoV-2 3CL protease in complex with compound 1 | | Descriptor: | 2-[4-[[4-[bis(fluoranyl)methoxy]-2-methyl-phenyl]amino]-2,6-bis(oxidanylidene)-3-[[3,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazin-1-yl]-N-methyl-ethanamide, 3C-like proteinase | | Authors: | Yamamoto, S, Tachibana, Y. | | Deposit date: | 2021-10-29 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of S-217622, a Noncovalent Oral SARS-CoV-2 3CL Protease Inhibitor Clinical Candidate for Treating COVID-19.
J.Med.Chem., 65, 2022
|
|
7EJY
 
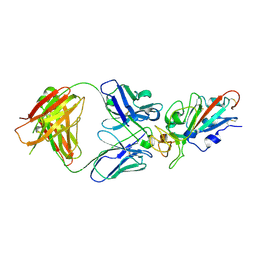 | | Complex Structure of antibody BD-503 and RBD of COVID-19 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain of BD-503, Light Chain of BD-503, ... | | Authors: | Xu, H, Wang, B, Zhao, T.N, Su, X.D. | | Deposit date: | 2021-04-03 | | Release date: | 2022-04-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Structure-based analyses of neutralization antibodies interacting with naturally occurring SARS-CoV-2 RBD variants.
Cell Res., 31, 2021
|
|
7EJZ
 
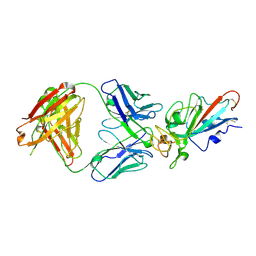 | | Complex Structure of antibody BD-503 and RBD-S477N of COVID-19 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain of BD-503, Light Chain of BD-503, ... | | Authors: | Xu, H, Wang, B, Zhao, T.N, Su, X.D. | | Deposit date: | 2021-04-03 | | Release date: | 2022-04-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.63 Å) | | Cite: | Structure-based analyses of neutralization antibodies interacting with naturally occurring SARS-CoV-2 RBD variants.
Cell Res., 31, 2021
|
|
7EK0
 
 | | Complex Structure of antibody BD-503 and RBD-N501Y of COVID-19 | | Descriptor: | Heavy Chain of BD-503, Light Chain of BD-503, Spike protein S1 | | Authors: | Xu, H, Wang, B, Zhao, T.N, Su, X.D. | | Deposit date: | 2021-04-03 | | Release date: | 2022-04-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-based analyses of neutralization antibodies interacting with naturally occurring SARS-CoV-2 RBD variants.
Cell Res., 31, 2021
|
|
7FJO
 
 | | Cryo-EM structure of South African (B.1.351) SARS-CoV-2 spike glycoprotein in complex with three T6 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, T6 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Zhang, S, Liang, Q. | | Deposit date: | 2021-08-04 | | Release date: | 2022-04-13 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | RBD trimer mRNA vaccine elicits broad and protective immune responses against SARS-CoV-2 variants.
Iscience, 25, 2022
|
|
7O35
 
 | |
7RAQ
 
 | |
7T01
 
 | | SARS-CoV-2 S-RBD + Fab 54042-4 | | Descriptor: | 54042-4 Fab - Heavy Chain, 54042-4 Fab - Light Chain, Spike protein S1 | | Authors: | Johnson, N.V, Mclellan, J.S. | | Deposit date: | 2021-11-29 | | Release date: | 2022-04-13 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Potent neutralization of SARS-CoV-2 variants of concern by an antibody with an uncommon genetic signature and structural mode of spike recognition.
Cell Rep, 37, 2021
|
|
7ULT
 
 | | Crystal Structure of SARS-CoV-2 Nsp16/10 Heterodimer Apo-Form. | | Descriptor: | 2'-O-methyltransferase, FORMIC ACID, Non-structural protein 10, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Rosas-Lemus, M, Kiryukhina, O, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-04-05 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of SARS-CoV-2 Nsp16/10 Heterodimer Apo-Form.
To Be Published
|
|
7WED
 
 | |
7WLC
 
 | |








