9BJ4
 
 | | Structure of the SARS-CoV-2 S 6P trimer complex with the human neutralizing antibody Fab fragment, C952 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C952 Heavy Chain, C952 Light Chain, ... | | Authors: | Rubio, A.A, Abernathy, M.E, Barnes, C.O. | | Deposit date: | 2024-04-24 | | Release date: | 2025-03-05 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Bispecific antibodies targeting the N-terminal and receptor binding domains potently neutralize SARS-CoV-2 variants of concern.
Sci Transl Med, 17, 2025
|
|
9CSS
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein Ecto-domain with internal tag, 1UP RBD conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Singh, S, Hasan, S.S. | | Deposit date: | 2024-07-24 | | Release date: | 2025-03-05 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Production and cryo-electron microscopy structure of an internally tagged SARS-CoV-2 spike ecto-domain construct.
J Struct Biol X, 11, 2025
|
|
9CT2
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein Ecto-domain with internal tag, All RBD down conformation, State-3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Singh, S, Hasan, S.S. | | Deposit date: | 2024-07-24 | | Release date: | 2025-03-05 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Production and cryo-electron microscopy structure of an internally tagged SARS-CoV-2 spike ecto-domain construct.
J Struct Biol X, 11, 2025
|
|
9CVH
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein Ecto-domain with internal tag, 1RBD UP, State-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(5-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Singh, S, Hasan, S.S. | | Deposit date: | 2024-07-29 | | Release date: | 2025-03-05 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Production and cryo-electron microscopy structure of an internally tagged SARS-CoV-2 spike ecto-domain construct.
J Struct Biol X, 11, 2025
|
|
9CXE
 
 | | SARS CoV-2 Spike protein Ectodomain with internal tag, all RBD-down conformation -C1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Singh, S, Hasan, S.S. | | Deposit date: | 2024-07-31 | | Release date: | 2025-03-05 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Production and cryo-electron microscopy structure of an internally tagged SARS-CoV-2 spike ecto-domain construct.
J Struct Biol X, 11, 2025
|
|
9HAJ
 
 | | Structure of compound 1 bound to SARS-CoV-2 main protease | | Descriptor: | (5~{R})-4-[(4-bromanyl-2-ethyl-phenyl)methyl]-1-(5-chloranylpyridin-3-yl)carbonyl-~{N}-ethyl-1,4-diazepane-5-carboxamide, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Mac Sweeney, A, Hazemann, J. | | Deposit date: | 2024-11-04 | | Release date: | 2025-03-05 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.276 Å) | | Cite: | Accelerating the Hit-To-Lead Optimization of a SARS-CoV-2 Mpro Inhibitor Series by Combining High-Throughput Medicinal Chemistry and Computational Simulations.
J.Med.Chem., 68, 2025
|
|
9I81
 
 | | SARS-CoV-2 RdRp bound to a stack of three HeE1-2Tyr molecules | | Descriptor: | N-[8-(cyclohexyloxy)-1-oxo-2-phenyl-1H-pyrido[2,1-b][1,3]benzothiazole-4-carbonyl]-L-tyrosine, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Kabinger, F, Doze, V, Schmitzova, J, Lidschreiber, M, Dienemann, C, Cramer, P. | | Deposit date: | 2025-02-04 | | Release date: | 2025-03-05 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural basis of SARS-CoV-2 polymerase inhibition by nonnucleoside inhibitor HeE1-2Tyr.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8U40
 
 | | Crystal structure of main protease of SARS-CoV-2 complexed with inhibitor | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-3-cyclopropyl-1-({(1E,2R)-1-imino-3-[(3R)-2-oxo-2,3-dihydropyridin-3-yl]propan-2-yl}amino)-1-oxopropan-2-yl]-5,7-difluoro-1H-indole-2-carboxamide | | Authors: | Chen, P, Arutyunova, E, Lu, J, Young, H.S, Lemieux, M.J. | | Deposit date: | 2023-09-08 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of main protease of SARS-CoV-2 complexed with inhibitor
To Be Published
|
|
8YK4
 
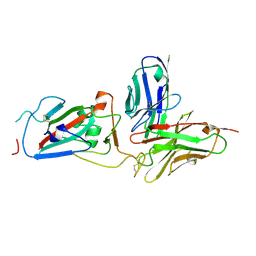 | | Structure of SARS-CoV-2 RBD and antibody NT-108 | | Descriptor: | Spike protein S1, antibody NT-108, single chain Fv fragment | | Authors: | Anraku, Y, Kita, S, Onodera, T, Sato, A, Tadokoro, T, Adachi, Y, Ito, S, Suzuki, T, Sasaki, J, Shiwa, N, Iwata, N, Nagata, N, Kazuki, Y, Oshimura, M, Sasaki, M, Orba, Y, Suzuki, T, Sawa, H, Hashiguchi, T, Fukuhara, H, Takahashi, Y, Maenaka, K. | | Deposit date: | 2024-03-04 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis for Potent Neutralization Activity of SARS-CoV-2 Antibody, NT-108
To Be Published
|
|
8YKG
 
 | | Structure of SARS-CoV-2 spike glycoprotein in complex with NT-108 scFv (1-up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NT-108 scFv, ... | | Authors: | Anraku, Y, Kita, S, Onodera, T, Tadokoro, T, Ito, S, Adachi, Y, Kotaki, R, Suzuki, T, Hashiguchi, T, Takahashi, Y, Maenaka, K. | | Deposit date: | 2024-03-05 | | Release date: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structural basis for potent neutralization activity of SARS-CoV-2 antibody, NT-108
To Be Published
|
|
8YKH
 
 | | Structure of SARS-CoV-2 spike RBD in complex with NT-108 scFv | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NT-108 scFv, Spike glycoprotein | | Authors: | Anraku, Y, Kita, S, Onodera, T, Tadokoro, T, Ito, S, Adachi, Y, Kotaki, R, Suzuki, T, Hashiguchi, T, Takahashi, Y, Maenaka, K. | | Deposit date: | 2024-03-05 | | Release date: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Structural basis for potent neutralization activity of SARS-CoV-2 antibody, NT-108
To Be Published
|
|
9BLL
 
 | | Cryo-EM of RBD(EG5.1)/1301B7 Fab Complex | | Descriptor: | 1301B7 Heavy Chain, 1301B7 Light Chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Walter, M.R, Green, T.J. | | Deposit date: | 2024-04-30 | | Release date: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Potent neutralization by a RBD antibody with broad specificity for SARS-CoV-2 JN.1 and other variants.
Npj Viruses, 2, 2024
|
|
9CCI
 
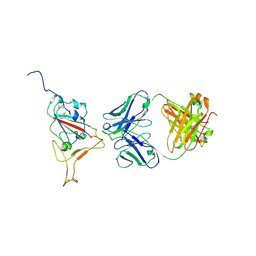 | |
9CCJ
 
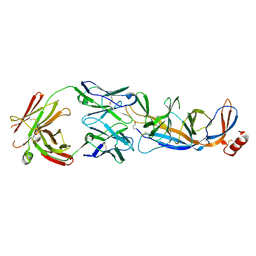 | |
9CGV
 
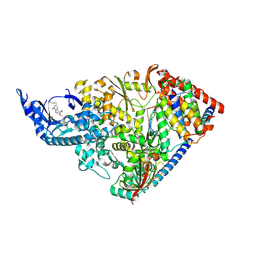 | | SARS-CoV-2 nsp12 NiRAN domain bound to a covalent inhibitor SW090466-1 | | Descriptor: | MANGANESE (II) ION, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Osinski, A, Hernandez, G, Tagliabracci, V.S. | | Deposit date: | 2024-07-01 | | Release date: | 2025-03-12 | | Last modified: | 2025-04-23 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Covalent inhibition of the SARS-CoV-2 NiRAN domain via an active-site cysteine.
J.Biol.Chem., 301, 2025
|
|
9J66
 
 | |
9N55
 
 | |
8UTC
 
 | | HUMAN LEUKOCYTE ANTIGEN B*07:02 IN COMPLEX WITH SARS-COV2 EPITOPE N105-113 (Y111F mutant) | | Descriptor: | Beta-2-microglobulin, CHLORIDE ION, HLA class I histocompatibility antigen, ... | | Authors: | Oltean, N, Nyovanie, S, Hashem, A, Patskovska, L, Patskovsky, Y, Krogsgaard, M. | | Deposit date: | 2023-10-30 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | HUMAN LEUKOCYTE ANTIGEN B*07:02 IN COMPLEX WITH SARS-COV2 EPITOPE N105-113 (Y111F mutant)
To Be Published
|
|
8Z1H
 
 | | Crystal structure of SARS main protease in complex with PF-00835231 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Lin, C, Zhang, J, Li, J. | | Deposit date: | 2024-04-11 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal structure of SARS main protease in complex with PF-00835231
To Be Published
|
|
9DJ8
 
 | | RNA-nsp9 bound to the NiRAN domain of the E-RTC with an empty G-pocket | | Descriptor: | ADENOSINE MONOPHOSPHATE, Non-structural protein 9, RNA-directed RNA polymerase, ... | | Authors: | Small, G.I, Darst, S.A, Campbell, E.A. | | Deposit date: | 2024-09-06 | | Release date: | 2025-03-19 | | Last modified: | 2025-10-01 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | The mechanism for GTP-mediated RNA capping by the SARS-CoV-2 NiRAN domain remains unresolved.
Cell, 188, 2025
|
|
9EL4
 
 | | Crystal Structure of SARS-CoV-2 Mpro mutant E166A with Pfizer Intravenous Inhibitor PF-00835231 | | Descriptor: | 3C-like proteinase nsp5, GLYCEROL, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Zvornicanin, S.N, Shaqra, A.M, Schiffer, C.A. | | Deposit date: | 2024-12-03 | | Release date: | 2025-03-19 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Molecular mechanisms of drug resistance and compensation in SARS-CoV-2 main protease: the interplay between E166 and L50.
Mbio, 16, 2025
|
|
9ELV
 
 | | Crystal Structure of SARS-CoV-2 Mpro mutant E166V with Pfizer Intravenous Inhibitor PF-00835231 | | Descriptor: | 3C-like proteinase nsp5, GLYCEROL, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Zvornicanin, S.N, Shaqra, A.M, Schiffer, C.A. | | Deposit date: | 2024-12-05 | | Release date: | 2025-03-19 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Molecular mechanisms of drug resistance and compensation in SARS-CoV-2 main protease: the interplay between E166 and L50.
Mbio, 16, 2025
|
|
9L09
 
 | | SARS-CoV-2 C-RTC with 13-TP | | Descriptor: | MAGNESIUM ION, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Huang, Y.C, Liang, L, Liu, Y.X, Yan, L.M, Lou, Z.Y, Rao, Z.H. | | Deposit date: | 2024-12-12 | | Release date: | 2025-03-19 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Discovery of a 2'-a-Fluoro-2'-b-C-(fluoromethyl) Purine Nucleotide Prodrug as a Potential Oral Anti-SARS-CoV-2 Agent
J Med Chem, 68, 2025
|
|
9L6C
 
 | | Cryo-EM structure of Delta RBD complexed with ConD-852, P2C-1F11 and S304 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab ConD-852, Heavy chain of Fab P2C-1F11, ... | | Authors: | Zheng, Z, Bing, J, Congcong, L, Bing, Z. | | Deposit date: | 2024-12-24 | | Release date: | 2025-03-19 | | Last modified: | 2025-10-01 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Structure and function of an unusual R452-dependent monoclonal antibody against SARS-CoV-2.
J.Virol., 99, 2025
|
|
9MEI
 
 | | Crystal Structure of SARS-CoV-2 Mpro mutant L50F E166V with Pfizer Intravenous Inhibitor PF-00835231 | | Descriptor: | 3C-like proteinase nsp5, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Zvornicanin, S.N, Shaqra, A.M, Schiffer, C.A. | | Deposit date: | 2024-12-06 | | Release date: | 2025-03-19 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Molecular mechanisms of drug resistance and compensation in SARS-CoV-2 main protease: the interplay between E166 and L50.
Mbio, 16, 2025
|
|








