7OD3
 
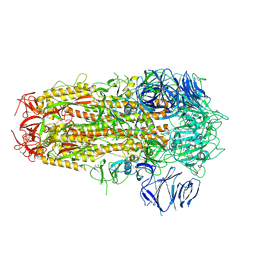 | | SARS CoV-2 Spike protein, Bristol UK Deletion variant, Closed conformation, C3 symmetry | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, Spike glycoprotein | | 著者 | Toelzer, C, Gupta, K, Yadav, S.K.N, Borucu, U, Schaffitzel, C, Berger, I. | | 登録日 | 2021-04-28 | | 公開日 | 2022-01-26 | | 最終更新日 | 2024-11-20 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structural insights in cell-type specific evolution of intra-host diversity by SARS-CoV-2.
Nat Commun, 13, 2022
|
|
7ODL
 
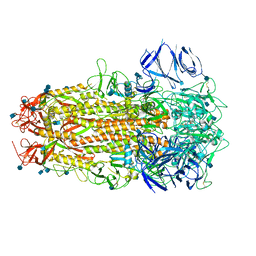 | | SARS CoV-2 Spike protein, Bristol UK Deletion variant, Closed conformation, C1 symmetry | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, Spike glycoprotein | | 著者 | Toelzer, C, Gupta, K, Yadav, S.K.N, Borucu, U, Schaffitzel, C, Berger, I. | | 登録日 | 2021-04-29 | | 公開日 | 2022-01-26 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (3.03 Å) | | 主引用文献 | Structural insights in cell-type specific evolution of intra-host diversity by SARS-CoV-2.
Nat Commun, 13, 2022
|
|
7QGI
 
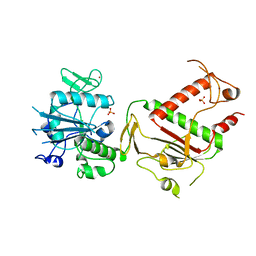 | | Crystal structure of SARS-CoV-2 NSP14 in the absence of NSP10 | | 分子名称: | PHOSPHATE ION, Proofreading exoribonuclease nsp14, ZINC ION | | 著者 | Newman, J.A, Imprachim, N, Yosaatmadja, Y, Gileadi, O. | | 登録日 | 2021-12-08 | | 公開日 | 2022-01-26 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Crystal structures and fragment screening of SARS-CoV-2 NSP14 reveal details of exoribonuclease activation and mRNA capping and provide starting points for antiviral drug development.
Nucleic Acids Res., 51, 2023
|
|
7QO9
 
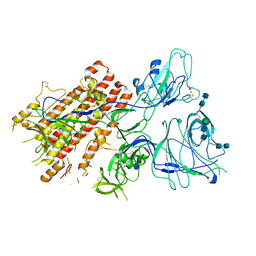 | | SARS-CoV-2 S Omicron Spike B.1.1.529 - RBD and NTD (Local) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,SARS-CoV-2 S Omicron Spike B.1.1.529, ... | | 著者 | Ni, D, Lau, K, Turelli, P, Beckert, B, Nazarov, S, Pojer, F, Myasnikov, A, Stahlberg, H, Trono, D. | | 登録日 | 2021-12-23 | | 公開日 | 2022-01-26 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (3.88 Å) | | 主引用文献 | Structural analysis of the Spike of the Omicron SARS-COV-2 variant by cryo-EM and implications for immune evasion
Biorxiv, 2021
|
|
7TI9
 
 | | Crystal structure of the ubiquitin-like domain 1 (Ubl1) of Nsp3 from SARS-CoV-2, form 2 | | 分子名称: | CHLORIDE ION, GLYCEROL, Papain-like protease nsp3 | | 著者 | Stogios, P.J, Skarina, T, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2022-01-13 | | 公開日 | 2022-01-26 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.73 Å) | | 主引用文献 | Crystal structure of the ubiquitin-like domain 1 (Ubl1) of Nsp3 from SARS-CoV-2, form 2
To Be Published
|
|
7TLL
 
 | | Structure of SARS-CoV-2 Mpro Omicron P132H in complex with Nirmatrelvir (PF-07321332) | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Greasley, S.E, Ferre, R.A, Plotnikova, O, Liu, W, Stewart, A.E. | | 登録日 | 2022-01-18 | | 公開日 | 2022-01-26 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Structural basis for the in vitro efficacy of nirmatrelvir against SARS-CoV-2 variants.
J.Biol.Chem., 298, 2022
|
|
7VJW
 
 | |
7VJX
 
 | |
7VJY
 
 | |
7VJZ
 
 | |
7VK0
 
 | |
7VK1
 
 | |
7VK2
 
 | |
7VK3
 
 | |
7VK4
 
 | |
7VK5
 
 | |
7VK6
 
 | |
7VK7
 
 | |
7VK8
 
 | |
7WEV
 
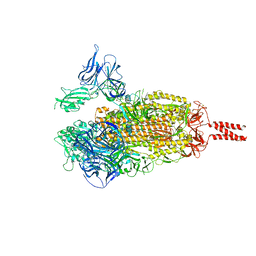 | |
7WK2
 
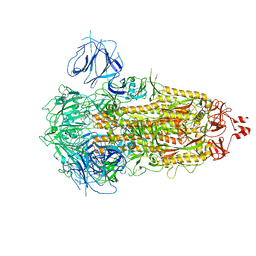 | | SARS-CoV-2 Omicron S-close | | 分子名称: | Spike glycoprotein | | 著者 | Li, J.W, Cong, Y. | | 登録日 | 2022-01-08 | | 公開日 | 2022-01-26 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Molecular basis of receptor binding and antibody neutralization of Omicron.
Nature, 604, 2022
|
|
7WK3
 
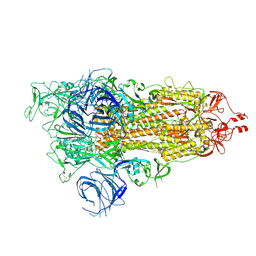 | | SARS-CoV-2 Omicron S-open | | 分子名称: | Spike glycoprotein | | 著者 | Li, J.W, Cong, Y. | | 登録日 | 2022-01-08 | | 公開日 | 2022-01-26 | | 最終更新日 | 2025-06-18 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural basis for ACE2 engagement and antibody evasion and neutralization of SARS-Co-2 Omicron varient
To Be Published
|
|
7WK8
 
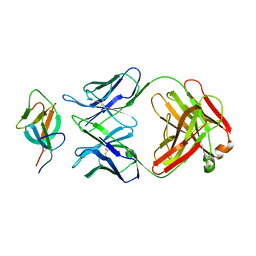 | |
7WKA
 
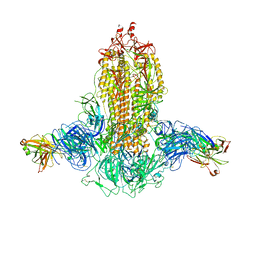 | |
7F7E
 
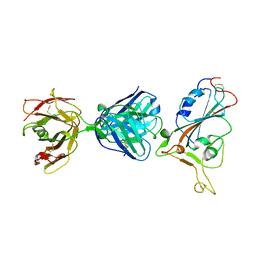 | | SARS-CoV-2 S protein RBD in complex with A5-10 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of A5-10 Fab, Light chain of A5-10 Fab, ... | | 著者 | Dou, Y, Wang, X, Wang, K, Liu, P, Lu, B. | | 登録日 | 2021-06-29 | | 公開日 | 2022-02-02 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.49 Å) | | 主引用文献 | Etesevimab in combination with JS026 neutralizing SARS-CoV-2 and its variants.
Emerg Microbes Infect, 11, 2022
|
|








