8F0H
 
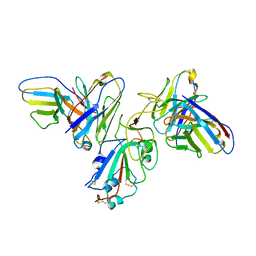 | | Structure of SARS-CoV-2 spike with antibody Fabs 2A10 and 1H2 (Local refinement of the RBD and Fabs 1H2 and 2A10) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody Fab 1H2 heavy chain, Antibody Fab 1H2 light chain, ... | | 著者 | Yu, X, Zyla, D, Hastie, K.M, Saphire, E.O. | | 登録日 | 2022-11-02 | | 公開日 | 2023-05-03 | | 実験手法 | ELECTRON MICROSCOPY (3.18 Å) | | 主引用文献 | Potent Omicron-neutralizing antibodies isolated from a patient vaccinated 6 months before Omicron emergence.
Cell Rep, 42, 2023
|
|
8GS9
 
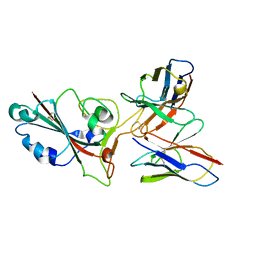 | | SARS-CoV-2 BA.2 spike RBD in complex bound with VacBB-551 | | 分子名称: | Heavy chain of VacBB-551, Light chain of VacBB-551, Spike glycoprotein | | 著者 | Liu, C.C, Ju, B, Shen, S.L, Zhang, Z. | | 登録日 | 2022-09-05 | | 公開日 | 2023-05-03 | | 最終更新日 | 2023-11-22 | | 実験手法 | ELECTRON MICROSCOPY (2.66 Å) | | 主引用文献 | Omicron BQ.1.1 and XBB.1 unprecedentedly escape broadly neutralizing antibodies elicited by prototype vaccination.
Cell Rep, 42, 2023
|
|
8OKK
 
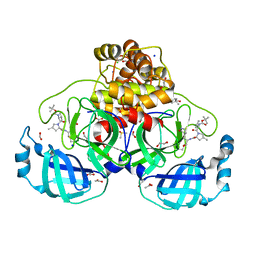 | | Crystal structure of F2F-2020184-00X bound to the main protease (3CLpro/Mpro) of SARS-CoV-2. | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ... | | 著者 | Costanzi, E, Demitri, N, Storici, P. | | 登録日 | 2023-03-28 | | 公開日 | 2023-05-03 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Broad-spectrum coronavirus 3C-like protease peptidomimetic inhibitors effectively block SARS-CoV-2 replication in cells: Design, synthesis, biological evaluation, and X-ray structure determination.
Eur.J.Med.Chem., 253, 2023
|
|
8OKL
 
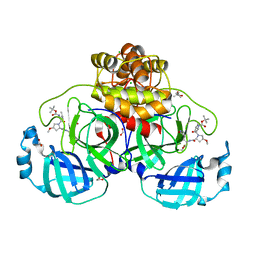 | | Crystal structure of F2F-2020185-01X bound to the main protease (3CLpro/Mpro) of SARS-CoV-2. | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ... | | 著者 | Costanzi, E, Demitri, N, Storici, P. | | 登録日 | 2023-03-28 | | 公開日 | 2023-05-03 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Broad-spectrum coronavirus 3C-like protease peptidomimetic inhibitors effectively block SARS-CoV-2 replication in cells: Design, synthesis, biological evaluation, and X-ray structure determination.
Eur.J.Med.Chem., 253, 2023
|
|
8OKM
 
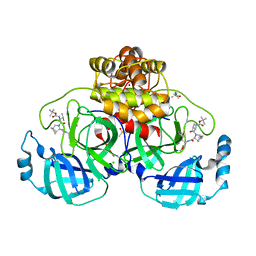 | | Crystal structure of F2F-2020197-00X bound to the main protease (3CLpro/Mpro) of SARS-CoV-2. | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ... | | 著者 | Costanzi, E, Demitri, N, Storici, P. | | 登録日 | 2023-03-28 | | 公開日 | 2023-05-03 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | Broad-spectrum coronavirus 3C-like protease peptidomimetic inhibitors effectively block SARS-CoV-2 replication in cells: Design, synthesis, biological evaluation, and X-ray structure determination.
Eur.J.Med.Chem., 253, 2023
|
|
8OKN
 
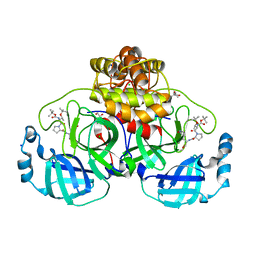 | | Crystal structure of F2F-2020198-00X bound to the main protease (3CLpro/Mpro) of SARS-CoV-2. | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ... | | 著者 | Costanzi, E, Demitri, N, Storici, P. | | 登録日 | 2023-03-28 | | 公開日 | 2023-05-03 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Broad-spectrum coronavirus 3C-like protease peptidomimetic inhibitors effectively block SARS-CoV-2 replication in cells: Design, synthesis, biological evaluation, and X-ray structure determination.
Eur.J.Med.Chem., 253, 2023
|
|
8OTO
 
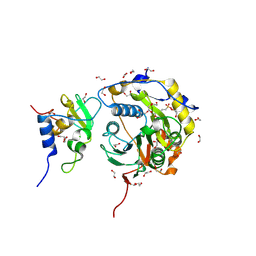 | |
7XSA
 
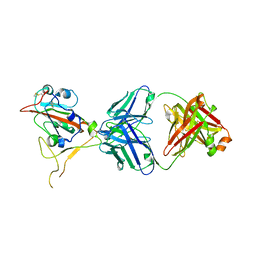 | |
7XSB
 
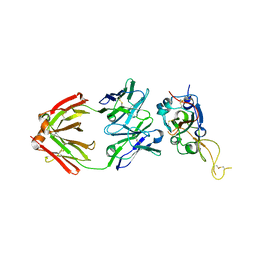 | |
8A99
 
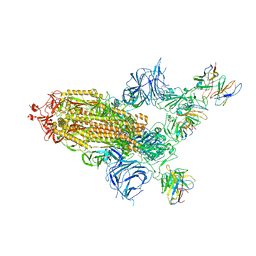 | | SARS Cov2 Spike in 1-up conformation complex with Fab47 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab47 (Heavy chain variable domain), ... | | 著者 | Hallberg, B.M, Das, H. | | 登録日 | 2022-06-28 | | 公開日 | 2023-05-10 | | 最終更新日 | 2023-09-27 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Immunoglobulin germline gene polymorphisms influence the function of SARS-CoV-2 neutralizing antibodies.
Immunity, 56, 2023
|
|
8H3M
 
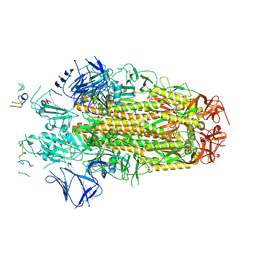 | | Conformation 1 of SARS-CoV-2 Omicron BA.1 Variant Spike protein complexed with MO1 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MO1 heavy chain, Spike glycoprotein | | 著者 | Ishimaru, H, Nishimura, M, Sutandhio, S, Shigematsu, H, Kato, K, Hasegawa, N, Mori, Y. | | 登録日 | 2022-10-09 | | 公開日 | 2023-05-10 | | 最終更新日 | 2023-08-02 | | 実験手法 | ELECTRON MICROSCOPY (2.48 Å) | | 主引用文献 | Identification and Analysis of Monoclonal Antibodies with Neutralizing Activity against Diverse SARS-CoV-2 Variants.
J.Virol., 97, 2023
|
|
8H3N
 
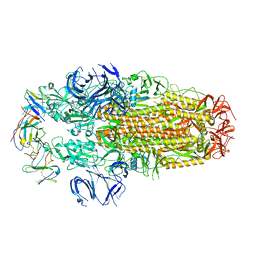 | | Conformation 2 of SARS-CoV-2 Omicron BA.1 Variant Spike protein complexed with MO1 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MO1 heavy-chain, MO1 light chain, ... | | 著者 | Ishimaru, H, Nishimura, M, Sutandhio, S, Shigematsu, H, Kato, K, Hasegawa, N, Mori, Y. | | 登録日 | 2022-10-09 | | 公開日 | 2023-05-10 | | 最終更新日 | 2023-08-02 | | 実験手法 | ELECTRON MICROSCOPY (2.73 Å) | | 主引用文献 | Identification and Analysis of Monoclonal Antibodies with Neutralizing Activity against Diverse SARS-CoV-2 Variants.
J.Virol., 97, 2023
|
|
8OT0
 
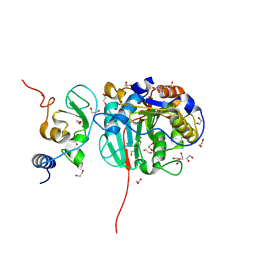 | | SARS-CoV-2 nsp10-16 methyltransferase in complex with MTA and glycine | | 分子名称: | 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | 著者 | Kremling, V, Sprenger, J, Oberthuer, D, Falke, S. | | 登録日 | 2023-04-20 | | 公開日 | 2023-05-10 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | Crystal structures of Tubercidin and Adenosine bound to the active site of the SARS-CoV-2 methyltransferase nsp10-16
To Be Published
|
|
8OV1
 
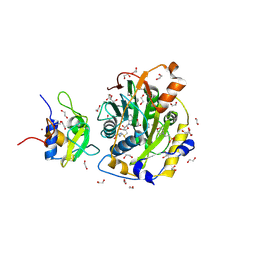 | |
8OV2
 
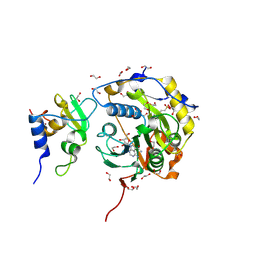 | |
8OV3
 
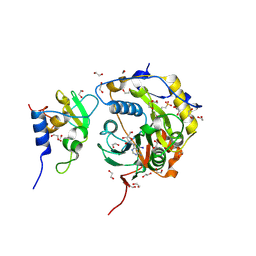 | | SARS-CoV-2 nsp10-16 methyltransferase in complex with 5-Iodotubercidin | | 分子名称: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, ... | | 著者 | Kremling, V, Sprenger, J, Oberthuer, D. | | 登録日 | 2023-04-25 | | 公開日 | 2023-05-10 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Crystal structures of Tubercidin and Adenosine bound to the active site of the SARS-CoV-2 methyltransferase nsp10-16
To Be Published
|
|
8OV4
 
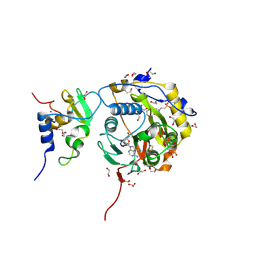 | |
8BSE
 
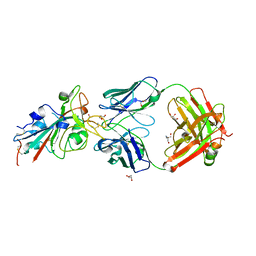 | | CRYSTAL STRUCTURE OF SARS-COV-2 RECEPTOR BINDING DOMAIN (RBD) in complex with 1D1 Fab | | 分子名称: | 1D1 FAB HEAVY CHAIN, 1D1 FAB LIGHT CHAIN, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | 著者 | Welin, M, Kimbung, Y.R, Focht, D, Pisitkun, T. | | 登録日 | 2022-11-25 | | 公開日 | 2023-05-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Efficacy of the combination of monoclonal antibodies against the SARS-CoV-2 Beta and Delta variants.
Plos One, 18, 2023
|
|
8BSF
 
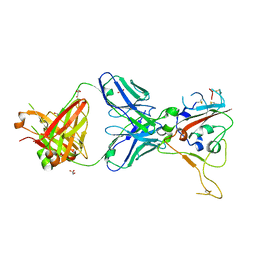 | | CRYSTAL STRUCTURE OF SARS-COV-2 RECEPTOR BINDING DOMAIN (RBD-beta variant) in complex with 3D2 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3D2 FAB HEAVY CHAIN, 3D2 FAB LIGHT CHAIN, ... | | 著者 | Welin, M, Kimbung, Y.R, Focht, D, Pisitkun, T. | | 登録日 | 2022-11-25 | | 公開日 | 2023-05-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Efficacy of the combination of monoclonal antibodies against the SARS-CoV-2 Beta and Delta variants.
Plos One, 18, 2023
|
|
8CSJ
 
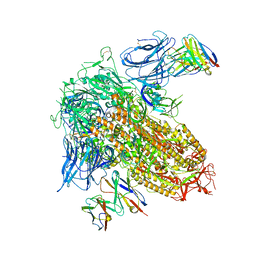 | |
8DK8
 
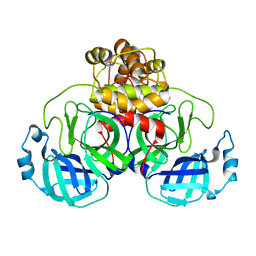 | |
8DL9
 
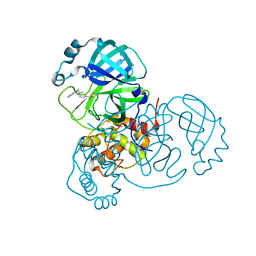 | |
8DLB
 
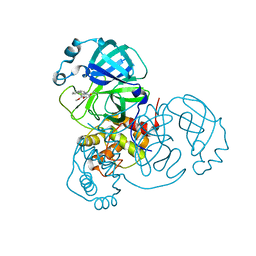 | | Room temperature X-ray structure of SARS-CoV-2 main protease in complex with compound Z2799209083 | | 分子名称: | 1-[(5S)-5-(3,4-dimethoxyphenyl)-3-phenyl-4,5-dihydro-1H-pyrazol-1-yl]ethan-1-one, 3C-like proteinase | | 著者 | Kovalevsky, A.Y, Coates, L, Kneller, D.W. | | 登録日 | 2022-07-07 | | 公開日 | 2023-05-17 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | AI-Accelerated Design of Targeted Covalent Inhibitors for SARS-CoV-2.
J.Chem.Inf.Model., 63, 2023
|
|
8DMD
 
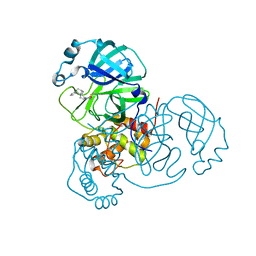 | |
8FWN
 
 | | Crystal structure of SARS-CoV-2 papain-like protease C111S mutant | | 分子名称: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | 著者 | Bezerra, E.H.S, Soprano, A.S, Tonoli, C.C.C, Prado, P.F.V, da Silva, J.C, Franchini, K.G, Trivella, D.B.B, Benedetti, C.E. | | 登録日 | 2023-01-23 | | 公開日 | 2023-05-17 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal structure of SARS-CoV-2 papain-like protease C111S mutant
To Be Published
|
|







