9ARU
 
 | | COVA2-15 fragment antigen binding in complex with SARS-CoV-2 6P-mut7 S protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COVA2-15 heavy chain variable region, ... | | Authors: | Ozorowski, G, Turner, H.L, Ward, A.B. | | Deposit date: | 2024-02-23 | | Release date: | 2025-01-29 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Plant-produced SARS-CoV-2 antibody engineered towards enhanced potency and in vivo efficacy.
Plant Biotechnol J, 23, 2025
|
|
9F2X
 
 | | Crystal structure of SARS-CoV-2 Mpro in complex with RHTCR03 | | Descriptor: | 3C-like proteinase nsp5, ~{tert}-butyl ~{N}-[4-[(2~{S})-1-[[(2~{S},3~{R})-4-azanyl-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]amino]-3-cyclopropyl-1-oxidanylidene-propan-2-yl]-3-oxidanylidene-pyrazin-2-yl]carbamate | | Authors: | El kilani, H, Hilgenfeld, R. | | Deposit date: | 2024-04-24 | | Release date: | 2025-01-29 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Based Optimization of Pyridone alpha-Ketoamides as Inhibitors of the SARS-CoV-2 Main Protease.
J.Med.Chem., 68, 2025
|
|
9F39
 
 | | Crystal structure of SARS-CoV-2 Mpro in complex with RK-54 | | Descriptor: | (2R,3R)-3-[[(2S)-3-cyclopropyl-2-[3-(2-methylpropanoylamino)-2-oxidanylidene-pyridin-1-yl]propanoyl]amino]-N-methyl-2-oxidanyl-4-[(3S)-2-oxidanylidenepyrrolidin-3-yl]butanamide, 3C-like proteinase nsp5 | | Authors: | El kilani, H, Hilgenfeld, R. | | Deposit date: | 2024-04-25 | | Release date: | 2025-01-29 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure-Based Optimization of Pyridone alpha-Ketoamides as Inhibitors of the SARS-CoV-2 Main Protease.
J.Med.Chem., 68, 2025
|
|
9F3A
 
 | | Crystal structure of SARS-CoV-2 Mpro in complex with RK-325 | | Descriptor: | 3C-like proteinase nsp5, tert-butyl N-[1-[(2S)-3-cyclopropyl-1-[[(2S,3R)-4-(methylamino)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]amino]-1-oxidanylidene-propan-2-yl]-5-fluoranyl-2-oxidanylidene-pyridin-3-yl]carbamate | | Authors: | El kilani, H, Hilgenfeld, R. | | Deposit date: | 2024-04-25 | | Release date: | 2025-01-29 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-Based Optimization of Pyridone alpha-Ketoamides as Inhibitors of the SARS-CoV-2 Main Protease.
J.Med.Chem., 68, 2025
|
|
9FHQ
 
 | | Crystal structure of SARS-CoV-2 Mpro in complex with RHTCR04 | | Descriptor: | 3C-like proteinase nsp5, ~{tert}-butyl ~{N}-[1-[(2~{S})-1-[[(2~{S})-4-azanyl-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]amino]-3-cyclopropyl-1-oxidanylidene-propan-2-yl]-6-oxidanylidene-pyrimidin-5-yl]carbamate | | Authors: | El kilani, H, Hilgenfeld, R. | | Deposit date: | 2024-05-28 | | Release date: | 2025-01-29 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Structure-Based Optimization of Pyridone alpha-Ketoamides as Inhibitors of the SARS-CoV-2 Main Protease.
J.Med.Chem., 68, 2025
|
|
9G9F
 
 | | CryoEM structure of Enterococcus italicus Csm-crRNA-CTR complex bound to AMPNPP | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, CRISPR system Cms endoribonuclease Csm3, CRISPR system Cms protein Csm2, ... | | Authors: | Jungfer, K, Jinek, M. | | Deposit date: | 2024-07-25 | | Release date: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Mechanistic determinants and dynamics of cA6 synthesis in type III CRISPR-Cas effector complexes.
Nucleic Acids Res., 53, 2025
|
|
9G9G
 
 | |
9G9H
 
 | |
9G9I
 
 | |
9G9K
 
 | |
9GMQ
 
 | | Crystal structure of the Mpro of SARS COV-2 in complex with the MG-87 inhibitor | | Descriptor: | 3C-like proteinase nsp5, CHLORIDE ION, ~{tert}-butyl ~{N}-[1-[(2~{S})-1-[[(2~{S},3~{R})-4-azanyl-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]amino]-3-cyclopropyl-1-oxidanylidene-propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | Authors: | El Kilani, H, Hilgenfeld, R. | | Deposit date: | 2024-08-29 | | Release date: | 2025-01-29 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structure-Based Optimization of Pyridone alpha-Ketoamides as Inhibitors of the SARS-CoV-2 Main Protease.
J.Med.Chem., 68, 2025
|
|
9GV2
 
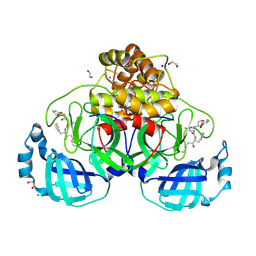 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the covalently bound inhibitor FP237 (compound 8p in publication) | | Descriptor: | (2S)-2-[2-(3-methoxyphenoxy)ethanoylamino]-4-methyl-N-[(2S)-3-oxidanylidene-1-phenyl-pentan-2-yl]pentanamide, 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ... | | Authors: | Strater, N, Sylvester, K, Muller, C.E, Flury, P, Kruger, N, Breidenbach, J, Guetschow, M, Laufer, S.A, Pillaiyar, T. | | Deposit date: | 2024-09-20 | | Release date: | 2025-01-29 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Design, Synthesis, and Unprecedented Interactions of Covalent Dipeptide-Based Inhibitors of SARS-CoV-2 Main Protease and Its Variants Displaying Potent Antiviral Activity.
J.Med.Chem., 68, 2025
|
|
9KT3
 
 | |
8VUO
 
 | |
8Y7T
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with C2 | | Descriptor: | 3C-like proteinase nsp5, 6-(iminomethyl)-4-(2-pyridin-2-ylethyl)-2-[4-(trifluoromethyl)phenyl]-1,2,4-triazine-3,5-dione | | Authors: | Zeng, R, Deng, X.Y, Yang, Z.Y, Wang, K, Jiang, Y.Y, Lei, J. | | Deposit date: | 2024-02-05 | | Release date: | 2025-02-05 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A deep learning model for structure-based bioactivity optimization and its application in the bioactivity optimization of a SARS-CoV-2 main protease inhibitor.
Eur.J.Med.Chem., 291, 2025
|
|
8Y7U
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with C5 | | Descriptor: | 2-(3-fluoro-4-(trifluoromethyl)phenyl)-6-(iminomethyl)-4-(2-oxo-2-(pyridin-2-yl)ethyl)-1,2,4-triazine-3,5(2H,4H)-dione, 3C-like proteinase nsp5 | | Authors: | Zeng, R, Deng, X.Y, Yang, Z.Y, Wang, K, Jiang, Y.Y, Lei, J. | | Deposit date: | 2024-02-05 | | Release date: | 2025-02-05 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A deep learning model for structure-based bioactivity optimization and its application in the bioactivity optimization of a SARS-CoV-2 main protease inhibitor.
Eur.J.Med.Chem., 291, 2025
|
|
8Z3W
 
 | |
8Z4X
 
 | |
8Z64
 
 | |
8Z6A
 
 | |
8Z7B
 
 | |
8Z7G
 
 | |
8Z7L
 
 | |
8Z7P
 
 | |
9CDK
 
 | | SARS-CoV-2 Mpro A173V mutant in complex with small molecule inhibitor Mpro61 | | Descriptor: | (5P)-5-[(1P,3M,3'P)-3-{3-chloro-5-[(2-chlorophenyl)methoxy]-4-fluorophenyl}-2-oxo-2H-[1,3'-bipyridin]-5-yl]-1-methylpyrimidine-2,4(1H,3H)-dione, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Tang, S, Kenneson, J.R, Anderson, K.S. | | Deposit date: | 2024-06-25 | | Release date: | 2025-02-05 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Exploring Possible Drug-Resistant Variants of SARS-CoV-2 Main Protease (M pro ) with Noncovalent Preclinical Candidate, Mpro61.
Acs Bio Med Chem Au, 5, 2025
|
|








