[English] 日本語
 Yorodumi
Yorodumi- PDB-6xvf: Crystal structure of bovine cytochrome bc1 in complex with tetrah... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6xvf | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Crystal structure of bovine cytochrome bc1 in complex with tetrahydro-quinolone inhibitor JAG021 | |||||||||
 Components Components |
| |||||||||
 Keywords Keywords | MEMBRANE PROTEIN / Cytochrome bc1 / Malaria / Electron transport | |||||||||
| Function / homology |  Function and homology information Function and homology informationComplex III assembly / subthalamus development / pons development / cerebellar Purkinje cell layer development / Respiratory electron transport / pyramidal neuron development / thalamus development / respiratory chain complex III / quinol-cytochrome-c reductase / quinol-cytochrome-c reductase activity ...Complex III assembly / subthalamus development / pons development / cerebellar Purkinje cell layer development / Respiratory electron transport / pyramidal neuron development / thalamus development / respiratory chain complex III / quinol-cytochrome-c reductase / quinol-cytochrome-c reductase activity / mitochondrial electron transport, ubiquinol to cytochrome c / Mitochondrial protein degradation / hypothalamus development / midbrain development / ubiquinone binding / respiratory electron transport chain / hippocampus development / metalloendopeptidase activity / 2 iron, 2 sulfur cluster binding / mitochondrial membrane / oxidoreductase activity / mitochondrial inner membrane / heme binding / mitochondrion / proteolysis / metal ion binding / membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 3.5 Å MOLECULAR REPLACEMENT / Resolution: 3.5 Å | |||||||||
 Authors Authors | Amporndanai, K. / Hasnain, S.S. / Antonyuk, S.V. | |||||||||
 Citation Citation |  Journal: Front Cell Infect Microbiol / Year: 2020 Journal: Front Cell Infect Microbiol / Year: 2020Title: Potent Tetrahydroquinolone Eliminates Apicomplexan Parasites. Authors: Martin J McPhillie / Ying Zhou / Mark R Hickman / James A Gordon / Christopher R Weber / Qigui Li / Patty J Lee / Kangsa Amporndanai / Rachel M Johnson / Heather Darby / Stuart Woods / Zhu- ...Authors: Martin J McPhillie / Ying Zhou / Mark R Hickman / James A Gordon / Christopher R Weber / Qigui Li / Patty J Lee / Kangsa Amporndanai / Rachel M Johnson / Heather Darby / Stuart Woods / Zhu-Hong Li / Richard S Priestley / Kurt D Ristroph / Scott B Biering / Kamal El Bissati / Seungmin Hwang / Farida Esaa Hakim / Sarah M Dovgin / Joseph D Lykins / Lucy Roberts / Kerrie Hargrave / Hua Cong / Anthony P Sinai / Stephen P Muench / Jitender P Dubey / Robert K Prud'homme / Hernan A Lorenzi / Giancarlo A Biagini / Silvia N Moreno / Craig W Roberts / Svetlana V Antonyuk / Colin W G Fishwick / Rima McLeod /   Abstract: Apicomplexan infections cause substantial morbidity and mortality, worldwide. New, improved therapies are needed. Herein, we create a next generation anti-apicomplexan lead compound, JAG21, a ...Apicomplexan infections cause substantial morbidity and mortality, worldwide. New, improved therapies are needed. Herein, we create a next generation anti-apicomplexan lead compound, JAG21, a tetrahydroquinolone, with increased sp3-character to improve parasite selectivity. Relative to other cytochrome inhibitors, JAG21 has improved solubility and ADMET properties, without need for pro-drug. JAG21 significantly reduces tachyzoites and encysted bradyzoites , and in primary and established chronic murine infections. Moreover, JAG21 treatment leads to 100% survival. Further, JAG21 is efficacious against drug-resistant . Causal prophylaxis and radical cure are achieved after sporozoite infection with oral administration of a single dose (2.5 mg/kg) or 3 days treatment at reduced dose (0.625 mg/kg/day), eliminating parasitemia, and leading to 100% survival. Enzymatic, binding, and co-crystallography/pharmacophore studies demonstrate selectivity for apicomplexan relative to mammalian enzymes. JAG21 has significant promise as a pre-clinical candidate for prevention, treatment, and cure of toxoplasmosis and malaria. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6xvf.cif.gz 6xvf.cif.gz | 842.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6xvf.ent.gz pdb6xvf.ent.gz | 688.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6xvf.json.gz 6xvf.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/xv/6xvf https://data.pdbj.org/pub/pdb/validation_reports/xv/6xvf ftp://data.pdbj.org/pub/pdb/validation_reports/xv/6xvf ftp://data.pdbj.org/pub/pdb/validation_reports/xv/6xvf | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  5okdS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| Unit cell |
|
- Components
Components
-Cytochrome b-c1 complex subunit ... , 8 types, 8 molecules ABEFGHIJ
| #1: Protein | Mass: 49165.152 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
|---|---|
| #2: Protein | Mass: 44495.000 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #5: Protein | Mass: 21640.580 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #6: Protein | Mass: 12485.247 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #7: Protein | Mass: 8799.227 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #8: Protein | Mass: 7515.433 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #9: Protein/peptide | Mass: 4827.567 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #10: Protein | Mass: 6866.897 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
-Protein , 2 types, 2 molecules CD
| #3: Protein | Mass: 42489.145 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
|---|---|
| #4: Protein | Mass: 27107.021 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
-Sugars , 1 types, 1 molecules 
| #16: Sugar | ChemComp-LMT / |
|---|
-Non-polymers , 11 types, 44 molecules 





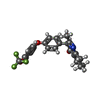














| #11: Chemical | | #12: Chemical | ChemComp-6PE / | #13: Chemical | ChemComp-PO4 / #14: Chemical | ChemComp-CDL / #15: Chemical | #17: Chemical | #18: Chemical | ChemComp-JAG / | #19: Chemical | ChemComp-HEC / | #20: Chemical | ChemComp-FES / | #21: Chemical | ChemComp-PX4 / | #22: Water | ChemComp-HOH / | |
|---|
-Details
| Has ligand of interest | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 4.86 Å3/Da / Density % sol: 74.7 % / Description: Bipyramidal red crystals |
|---|---|
| Crystal grow | Temperature: 277 K / Method: vapor diffusion, hanging drop / pH: 6.8 Details: Protein 40mg/mL with 1.6% HECAMEG; Reservoir solution: 50mM KPi pH 6.8, 100mM NaCl, 3mM NaN3, 10-13% PEG4000 |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SOLEIL SOLEIL  / Beamline: PROXIMA 2 / Wavelength: 0.9801 Å / Beamline: PROXIMA 2 / Wavelength: 0.9801 Å |
| Detector | Type: DECTRIS EIGER2 X 9M / Detector: PIXEL / Date: Dec 1, 2017 / Details: mirrors |
| Radiation | Monochromator: Si111 / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.9801 Å / Relative weight: 1 |
| Reflection | Resolution: 3.45→90.88 Å / Num. obs: 53526 / % possible obs: 91.4 % / Redundancy: 11.4 % / Biso Wilson estimate: 94.4 Å2 / CC1/2: 0.999 / Rmerge(I) obs: 0.225 / Rpim(I) all: 0.068 / Rrim(I) all: 0.235 / Net I/σ(I): 8.6 |
| Reflection shell | Resolution: 3.45→3.56 Å / Redundancy: 11.6 % / Rmerge(I) obs: 1.111 / Mean I/σ(I) obs: 2.4 / Num. unique obs: 4659 / CC1/2: 0.295 / Rpim(I) all: 0.334 / % possible all: 92.8 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 5OKD Resolution: 3.5→90.88 Å / Cor.coef. Fo:Fc: 0.874 / Cor.coef. Fo:Fc free: 0.911 / SU B: 0.584 / SU ML: 0.374 / Cross valid method: THROUGHOUT / σ(F): 0 / ESU R Free: 0.493 / Details: U VALUES : WITH TLS ADDED
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 304.39 Å2 / Biso mean: 165.129 Å2 / Biso min: 28.77 Å2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 3.5→90.88 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 3.5→3.591 Å / Rfactor Rfree error: 0
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller













 PDBj
PDBj


























