[English] 日本語
 Yorodumi
Yorodumi- PDB-5okd: Crystal structure of bovine Cytochrome bc1 in complex with inhibi... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 5okd | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Crystal structure of bovine Cytochrome bc1 in complex with inhibitor SCR0911. | |||||||||
 Components Components |
| |||||||||
 Keywords Keywords | ELECTRON TRANSPORT / antimalarial inhibitor / Qi site binder | |||||||||
| Function / homology |  Function and homology information Function and homology informationComplex III assembly / subthalamus development / pons development / cerebellar Purkinje cell layer development / Respiratory electron transport / pyramidal neuron development / thalamus development / respiratory chain complex III / quinol-cytochrome-c reductase / quinol-cytochrome-c reductase activity ...Complex III assembly / subthalamus development / pons development / cerebellar Purkinje cell layer development / Respiratory electron transport / pyramidal neuron development / thalamus development / respiratory chain complex III / quinol-cytochrome-c reductase / quinol-cytochrome-c reductase activity / mitochondrial electron transport, ubiquinol to cytochrome c / Mitochondrial protein degradation / hypothalamus development / midbrain development / ubiquinone binding / respiratory electron transport chain / hippocampus development / metalloendopeptidase activity / 2 iron, 2 sulfur cluster binding / mitochondrial membrane / oxidoreductase activity / mitochondrial inner membrane / heme binding / mitochondrion / proteolysis / metal ion binding / membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 3.1 Å MOLECULAR REPLACEMENT / Resolution: 3.1 Å | |||||||||
 Authors Authors | Amporndanai, K. / O'Neill, P.M. / Hasnain, S.S. / Antonyuk, S.V. | |||||||||
 Citation Citation |  Journal: IUCrJ / Year: 2018 Journal: IUCrJ / Year: 2018Title: X-ray and cryo-EM structures of inhibitor-bound cytochrome complexes for structure-based drug discovery. Authors: Kangsa Amporndanai / Rachel M Johnson / Paul M O'Neill / Colin W G Fishwick / Alexander H Jamson / Shaun Rawson / Stephen P Muench / S Samar Hasnain / Svetlana V Antonyuk /  Abstract: Cytochrome , a dimeric multi-subunit electron-transport protein embedded in the inner mitochondrial membrane, is a major drug target for the treatment and prevention of malaria and toxoplasmosis. ...Cytochrome , a dimeric multi-subunit electron-transport protein embedded in the inner mitochondrial membrane, is a major drug target for the treatment and prevention of malaria and toxoplasmosis. Structural studies of cytochrome from mammalian homologues co-crystallized with lead compounds have underpinned structure-based drug design to develop compounds with higher potency and selectivity. However, owing to the limited amount of cytochrome that may be available from parasites, all efforts have been focused on homologous cytochrome complexes from mammalian species, which has resulted in the failure of some drug candidates owing to toxicity in the host. Crystallographic studies of the native parasite proteins are not feasible owing to limited availability of the proteins. Here, it is demonstrated that cytochrome is highly amenable to single-particle cryo-EM (which uses significantly less protein) by solving the apo and two inhibitor-bound structures to ∼4.1 Å resolution, revealing clear inhibitor density at the binding site. Therefore, cryo-EM is proposed as a viable alternative method for structure-based drug discovery using both host and parasite enzymes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  5okd.cif.gz 5okd.cif.gz | 861 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb5okd.ent.gz pdb5okd.ent.gz | 699.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  5okd.json.gz 5okd.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ok/5okd https://data.pdbj.org/pub/pdb/validation_reports/ok/5okd ftp://data.pdbj.org/pub/pdb/validation_reports/ok/5okd ftp://data.pdbj.org/pub/pdb/validation_reports/ok/5okd | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  4286C  4288C  4292C  6fo0C  6fo2C  6fo6C  4d6tS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
| ||||||||
| Components on special symmetry positions |
|
- Components
Components
-Cytochrome b-c1 complex subunit ... , 8 types, 8 molecules ABEFGHIJ
| #1: Protein | Mass: 52840.461 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
|---|---|
| #2: Protein | Mass: 48203.434 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #5: Protein | Mass: 21640.580 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #6: Protein | Mass: 13502.387 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #7: Protein | Mass: 9737.223 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #8: Protein | Mass: 10638.744 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #9: Protein/peptide | Mass: 4898.645 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #10: Protein | Mass: 7469.621 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
-Protein , 2 types, 2 molecules CD
| #3: Protein | Mass: 42620.340 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
|---|---|
| #4: Protein | Mass: 35343.770 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
-Sugars , 1 types, 1 molecules 
| #16: Sugar | ChemComp-LMT / |
|---|
-Non-polymers , 11 types, 104 molecules 



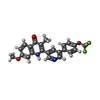
















| #11: Chemical | | #12: Chemical | ChemComp-6PE / | #13: Chemical | ChemComp-CDL / #14: Chemical | #15: Chemical | ChemComp-9XE / | #17: Chemical | #18: Chemical | ChemComp-HEC / | #19: Chemical | ChemComp-PO4 / #20: Chemical | ChemComp-FES / | #21: Chemical | ChemComp-PX4 / | #22: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 4.41 Å3/Da / Density % sol: 72.08 % / Description: Bipyramidal red |
|---|---|
| Crystal grow | Temperature: 277 K / Method: vapor diffusion, hanging drop Details: Protein 40mg/mL with 1.6% HECAMEG; reservoir solution 50mM KPi pH 6.8, 100mM NaCl, 3mM NaN3, 10-13% PEG4000 |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  Diamond Diamond  / Beamline: I03 / Wavelength: 1 Å / Beamline: I03 / Wavelength: 1 Å |
| Detector | Type: DECTRIS PILATUS 6M / Detector: PIXEL / Date: Feb 27, 2017 / Details: mirrors |
| Radiation | Monochromator: Si111 / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1 Å / Relative weight: 1 |
| Reflection | Resolution: 3.1→89 Å / Num. obs: 80268 / % possible obs: 99.5 % / Redundancy: 7.4 % / Biso Wilson estimate: 71.303 Å2 / CC1/2: 0.977 / Rmerge(I) obs: 0.104 / Rpim(I) all: 0.043 / Net I/σ(I): 12.3 |
| Reflection shell | Resolution: 3.1→3.16 Å / Redundancy: 7.5 % / Rmerge(I) obs: 1.126 / Mean I/σ(I) obs: 1.8 / Num. unique obs: 4428 / CC1/2: 0.572 / Rpim(I) all: 0.655 / % possible all: 99.9 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 4d6t Resolution: 3.1→31.76 Å / Cor.coef. Fo:Fc: 0.936 / Cor.coef. Fo:Fc free: 0.915 / SU B: 37.897 / SU ML: 0.295 / Cross valid method: THROUGHOUT / ESU R: 0.821 / ESU R Free: 0.353 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å / Solvent model: MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 101.1 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: 1 / Resolution: 3.1→31.76 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj

























