+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4288 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of bovine cytochrome bc1 with no ligand bound | |||||||||
 Map data Map data | The cryoEM map of bovine bc1 with no ligand bound. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Cryo-EM / Inhibitor binding / Membrane protein / cytochrome bc1 | |||||||||
| Function / homology |  Function and homology information Function and homology informationComplex III assembly / subthalamus development / pons development / cerebellar Purkinje cell layer development / Respiratory electron transport / pyramidal neuron development / thalamus development / respiratory chain complex III / quinol-cytochrome-c reductase / quinol-cytochrome-c reductase activity ...Complex III assembly / subthalamus development / pons development / cerebellar Purkinje cell layer development / Respiratory electron transport / pyramidal neuron development / thalamus development / respiratory chain complex III / quinol-cytochrome-c reductase / quinol-cytochrome-c reductase activity / mitochondrial electron transport, ubiquinol to cytochrome c / Mitochondrial protein degradation / hypothalamus development / midbrain development / ubiquinone binding / respiratory electron transport chain / hippocampus development / metalloendopeptidase activity / 2 iron, 2 sulfur cluster binding / mitochondrial membrane / oxidoreductase activity / mitochondrial inner membrane / heme binding / mitochondrion / proteolysis / metal ion binding / membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
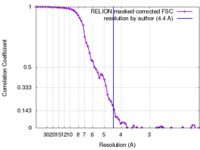
| Method | single particle reconstruction / cryo EM / Resolution: 4.4 Å | |||||||||
 Authors Authors | Johnson RM / Amporndanai K | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: IUCrJ / Year: 2018 Journal: IUCrJ / Year: 2018Title: X-ray and cryo-EM structures of inhibitor-bound cytochrome complexes for structure-based drug discovery. Authors: Kangsa Amporndanai / Rachel M Johnson / Paul M O'Neill / Colin W G Fishwick / Alexander H Jamson / Shaun Rawson / Stephen P Muench / S Samar Hasnain / Svetlana V Antonyuk /  Abstract: Cytochrome , a dimeric multi-subunit electron-transport protein embedded in the inner mitochondrial membrane, is a major drug target for the treatment and prevention of malaria and toxoplasmosis. ...Cytochrome , a dimeric multi-subunit electron-transport protein embedded in the inner mitochondrial membrane, is a major drug target for the treatment and prevention of malaria and toxoplasmosis. Structural studies of cytochrome from mammalian homologues co-crystallized with lead compounds have underpinned structure-based drug design to develop compounds with higher potency and selectivity. However, owing to the limited amount of cytochrome that may be available from parasites, all efforts have been focused on homologous cytochrome complexes from mammalian species, which has resulted in the failure of some drug candidates owing to toxicity in the host. Crystallographic studies of the native parasite proteins are not feasible owing to limited availability of the proteins. Here, it is demonstrated that cytochrome is highly amenable to single-particle cryo-EM (which uses significantly less protein) by solving the apo and two inhibitor-bound structures to ∼4.1 Å resolution, revealing clear inhibitor density at the binding site. Therefore, cryo-EM is proposed as a viable alternative method for structure-based drug discovery using both host and parasite enzymes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4288.map.gz emd_4288.map.gz | 28.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4288-v30.xml emd-4288-v30.xml emd-4288.xml emd-4288.xml | 30 KB 30 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4288_fsc.xml emd_4288_fsc.xml | 7.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_4288.png emd_4288.png | 46.1 KB | ||
| Filedesc metadata |  emd-4288.cif.gz emd-4288.cif.gz | 7.7 KB | ||
| Others |  emd_4288_additional.map.gz emd_4288_additional.map.gz emd_4288_half_map_1.map.gz emd_4288_half_map_1.map.gz emd_4288_half_map_2.map.gz emd_4288_half_map_2.map.gz | 22.4 MB 22.5 MB 22.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4288 http://ftp.pdbj.org/pub/emdb/structures/EMD-4288 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4288 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4288 | HTTPS FTP |
-Related structure data
| Related structure data |  6fo2MC  4286C  4292C  5okdC  6fo0C  6fo6C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_4288.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4288.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The cryoEM map of bovine bc1 with no ligand bound. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.047 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: The unsharpened map of apo-bc1.
| File | emd_4288_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The unsharpened map of apo-bc1. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: The first half map of apo-bc1.
| File | emd_4288_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The first half map of apo-bc1. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: The second half map of apo-bc1.
| File | emd_4288_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The second half map of apo-bc1. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : Bovine cytochrome bc1 in the absence of any inhibitor.
+Supramolecule #1: Bovine cytochrome bc1 in the absence of any inhibitor.
+Macromolecule #1: Cytochrome b-c1 complex subunit 1, mitochondrial
+Macromolecule #2: Cytochrome b-c1 complex subunit 2, mitochondrial
+Macromolecule #3: Cytochrome b
+Macromolecule #4: Cytochrome c1, heme protein, mitochondrial
+Macromolecule #5: Cytochrome b-c1 complex subunit Rieske, mitochondrial
+Macromolecule #6: Cytochrome b-c1 complex subunit 7
+Macromolecule #7: Cytochrome b-c1 complex subunit 8
+Macromolecule #8: Cytochrome b-c1 complex subunit 6, mitochondrial
+Macromolecule #9: Cytochrome b-c1 complex subunit Rieske, mitochondrial
+Macromolecule #10: Cytochrome b-c1 complex subunit 9
+Macromolecule #11: PROTOPORPHYRIN IX CONTAINING FE
+Macromolecule #12: HEME C
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5.0 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV Details: blot for 6 seconds with a blot force of 6 before plunge-freezing. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 3256 / Average exposure time: 12.0 sec. / Average electron dose: 44.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 4.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 79000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-6fo2: |
 Movie
Movie Controller
Controller



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)