[English] 日本語
 Yorodumi
Yorodumi- PDB-6t22: N-terminal domain of EcoKMcrA restriction endonuclease (NEco) in ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6t22 | ||||||
|---|---|---|---|---|---|---|---|
| Title | N-terminal domain of EcoKMcrA restriction endonuclease (NEco) in complex with T5hmCGA target sequence | ||||||
 Components Components |
| ||||||
 Keywords Keywords | HYDROLASE / EcoKMcrA / NEco / N-TERMINAL DOMAIN / MODIFICATION DEPENDENT RESTRICTION / 5-METHYLCYTOSINE / 5MC / 5-HYDROXYMETHYLCYTOSINE / 5HMC / HNH ENDONUCLEASE / BBA-ME NUCLEASE | ||||||
| Function / homology |  Function and homology information Function and homology informationHydrolases; Acting on ester bonds; Endodeoxyribonucleases producing 5'-phosphomonoesters / methyl-CpG binding / DNA restriction-modification system / endonuclease activity / zinc ion binding Similarity search - Function | ||||||
| Biological species |  synthetic construct (others) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.21 Å MOLECULAR REPLACEMENT / Resolution: 2.21 Å | ||||||
 Authors Authors | Slyvka, A. / Zagorskaite, E. / Czapinska, H. / Sasnauskas, G. / Bochtler, M. | ||||||
 Citation Citation |  Journal: Nucleic Acids Res. / Year: 2019 Journal: Nucleic Acids Res. / Year: 2019Title: Crystal structure of the EcoKMcrA N-terminal domain (NEco): recognition of modified cytosine bases without flipping. Authors: Slyvka, A. / Zagorskaite, E. / Czapinska, H. / Sasnauskas, G. / Bochtler, M. #1:  Journal: Nucleic Acids Res. / Year: 2018 Journal: Nucleic Acids Res. / Year: 2018Title: Activity and structure of EcoKMcrA. Authors: Czapinska, H. / Kowalska, M. / Zagorskaite, E. / Manakova, E. / Slyvka, A. / Xu, S.Y. / Siksnys, V. / Sasnauskas, G. / Bochtler, M. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6t22.cif.gz 6t22.cif.gz | 189.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6t22.ent.gz pdb6t22.ent.gz | 147.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6t22.json.gz 6t22.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/t2/6t22 https://data.pdbj.org/pub/pdb/validation_reports/t2/6t22 ftp://data.pdbj.org/pub/pdb/validation_reports/t2/6t22 ftp://data.pdbj.org/pub/pdb/validation_reports/t2/6t22 | HTTPS FTP |
|---|
-Related structure data
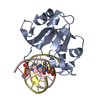
| Related structure data | 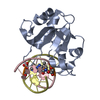 6r64C 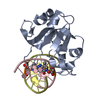 6t21C  6ghcS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
| ||||||||
| Components on special symmetry positions |
|
- Components
Components
| #1: Protein | Mass: 17368.604 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   References: UniProt: P24200, Hydrolases; Acting on ester bonds; Endodeoxyribonucleases producing 5'-phosphomonoesters #2: DNA chain | Mass: 3025.005 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.) synthetic construct (others) / Production host: synthetic construct (others) #3: DNA chain | Mass: 3123.080 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.) synthetic construct (others) / Production host: synthetic construct (others) #4: Water | ChemComp-HOH / | Has ligand of interest | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.09 Å3/Da / Density % sol: 60.15 % |
|---|---|
| Crystal grow | Temperature: 291 K / Method: vapor diffusion, hanging drop / pH: 6.5 Details: 1.8 ul of protein-DNA solution (436:523 uM) was mixed with 2.2 ul of the F1 condition of the PACT premier crystal screen (MDL) (0.2 M Sodium fluoride, 0.1 M Bis-Tris propane, pH 6.5, 20% PEG ...Details: 1.8 ul of protein-DNA solution (436:523 uM) was mixed with 2.2 ul of the F1 condition of the PACT premier crystal screen (MDL) (0.2 M Sodium fluoride, 0.1 M Bis-Tris propane, pH 6.5, 20% PEG 3350) and cryo-protected with an addition of 25% glycerol. |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  PETRA III, EMBL c/o DESY PETRA III, EMBL c/o DESY  / Beamline: P14 (MX2) / Wavelength: 0.9195 Å / Beamline: P14 (MX2) / Wavelength: 0.9195 Å |
| Detector | Type: DECTRIS EIGER X 16M / Detector: PIXEL / Date: Aug 14, 2019 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.9195 Å / Relative weight: 1 |
| Reflection | Resolution: 2.21→33.54 Å / Num. obs: 30298 / % possible obs: 99.2 % / Redundancy: 35.8 % / Biso Wilson estimate: 55.6 Å2 / CC1/2: 1 / Rmerge(I) obs: 0.084 / Rrim(I) all: 0.086 / Net I/σ(I): 38.15 |
| Reflection shell | Resolution: 2.21→2.34 Å / Redundancy: 20.8 % / Rmerge(I) obs: 1.761 / Mean I/σ(I) obs: 1.91 / Num. unique obs: 4597 / CC1/2: 0.734 / Rrim(I) all: 1.802 / % possible all: 95.4 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 6GHC Resolution: 2.21→33.54 Å / Cor.coef. Fo:Fc: 0.974 / Cor.coef. Fo:Fc free: 0.958 / SU B: 8.998 / SU ML: 0.117 / Cross valid method: THROUGHOUT / σ(F): 0 / ESU R: 0.18 / ESU R Free: 0.163 Details: HYDROGEN ATOMS HAVE BEEN ADDED IN THE RIDING POSITIONS. TLS REFINEMENT HAS BEEN USED. U VALUES : WITH TLS ADDED. THE CLUSTERS OF SOLVENT MOLECULES BETWEEN RESIDUES 46 AND 94 AND NEXT TO ...Details: HYDROGEN ATOMS HAVE BEEN ADDED IN THE RIDING POSITIONS. TLS REFINEMENT HAS BEEN USED. U VALUES : WITH TLS ADDED. THE CLUSTERS OF SOLVENT MOLECULES BETWEEN RESIDUES 46 AND 94 AND NEXT TO RESIDUE 54 OF CHAIN B LIKELY CORRESPOND TO DISORDERED GLYCEROL MOLECULES. THE ELECTRON DENSITY IS NOT DEFINED ENOUGH TO UNAMBIGUOUSLY MODEL THEM.
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 158.21 Å2 / Biso mean: 53.522 Å2 / Biso min: 35.08 Å2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 2.21→33.54 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.21→2.26 Å / Rfactor Rfree error: 0
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller










 PDBj
PDBj

