+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6jb3 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of SUR1 subunit bound with repaglinide | |||||||||||||||
 Components Components | ATP-binding cassette sub-family C member 8 isoform X2 | |||||||||||||||
 Keywords Keywords | MEMBRANE PROTEIN / KATP / channel / repaglinide / Kir / ABC transporter / SUR / diabetes / insulin secretagogue | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationglutamate secretion, neurotransmission / inward rectifying potassium channel / sulfonylurea receptor activity / ATPase-coupled monoatomic cation transmembrane transporter activity / : / neuromuscular process / action potential / potassium channel activity / ABC-type transporter activity / positive regulation of insulin secretion involved in cellular response to glucose stimulus ...glutamate secretion, neurotransmission / inward rectifying potassium channel / sulfonylurea receptor activity / ATPase-coupled monoatomic cation transmembrane transporter activity / : / neuromuscular process / action potential / potassium channel activity / ABC-type transporter activity / positive regulation of insulin secretion involved in cellular response to glucose stimulus / cellular response to nutrient levels / negative regulation of insulin secretion / ADP binding / sarcolemma / presynapse / transmembrane transporter binding / response to xenobiotic stimulus / ATP hydrolysis activity / ATP binding / metal ion binding Similarity search - Function | |||||||||||||||
| Biological species |  Mesocricetus auratus (golden hamster) Mesocricetus auratus (golden hamster) | |||||||||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 3.53 Å | |||||||||||||||
 Authors Authors | Chen, L. / Ding, D. / Wang, M. / Wu, J.-X. / Kang, Y. | |||||||||||||||
| Funding support |  China, 4items China, 4items
| |||||||||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2019 Journal: Cell Rep / Year: 2019Title: The Structural Basis for the Binding of Repaglinide to the Pancreatic K Channel. Authors: Dian Ding / Mengmeng Wang / Jing-Xiang Wu / Yunlu Kang / Lei Chen /  Abstract: Repaglinide (RPG) is a short-acting insulin secretagogue widely prescribed for the treatment of type 2 diabetes. It boosts insulin secretion by inhibiting the pancreatic ATP-sensitive potassium ...Repaglinide (RPG) is a short-acting insulin secretagogue widely prescribed for the treatment of type 2 diabetes. It boosts insulin secretion by inhibiting the pancreatic ATP-sensitive potassium channel (K). However, the mechanisms by which RPG binds to the K channel are poorly understood. Here, we describe two cryo-EM structures: the pancreatic K channel in complex with inhibitory RPG and adenosine-5'-(γ-thio)-triphosphate (ATPγS) at 3.3 Å and a medium-resolution structure of a RPG-bound mini SUR1 protein in which the N terminus of the inward-rectifying potassium channel 6.1 (Kir6.1) is fused to the ABC transporter module of the sulfonylurea receptor 1 (SUR1). These structures reveal the binding site of RPG in the SUR1 subunit. Furthermore, the high-resolution structure reveals the complex architecture of the ATP binding site, which is formed by both Kir6.2 and SUR1 subunits, and the domain-domain interaction interfaces. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6jb3.cif.gz 6jb3.cif.gz | 168.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6jb3.ent.gz pdb6jb3.ent.gz | 117.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6jb3.json.gz 6jb3.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/jb/6jb3 https://data.pdbj.org/pub/pdb/validation_reports/jb/6jb3 ftp://data.pdbj.org/pub/pdb/validation_reports/jb/6jb3 ftp://data.pdbj.org/pub/pdb/validation_reports/jb/6jb3 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  9788MC  9787C  9789C  6jb1C M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
| #1: Protein | Mass: 177295.516 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Mesocricetus auratus (golden hamster) / Gene: Abcc8 / Production host: Mesocricetus auratus (golden hamster) / Gene: Abcc8 / Production host:  Homo sapiens (human) / References: UniProt: A0A1U7R319 Homo sapiens (human) / References: UniProt: A0A1U7R319 |
|---|---|
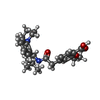
| #2: Chemical | ChemComp-BJX / |
| #3: Chemical | ChemComp-AJP / |
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: SUR1 / Type: COMPLEX / Entity ID: #1 / Source: RECOMBINANT |
|---|---|
| Source (natural) | Organism:  |
| Source (recombinant) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Buffer solution | pH: 7.5 |
| Specimen | Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD |
| Image recording | Electron dose: 50 e/Å2 / Film or detector model: GATAN K2 QUANTUM (4k x 4k) |
- Processing
Processing
| Software | Name: PHENIX / Version: (1.12_2829: ???) / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EM software | Name: RELION / Version: 2 / Category: 3D reconstruction | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CTF correction | Type: NONE | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3D reconstruction | Resolution: 3.53 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 653991 / Symmetry type: POINT | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | Resolution: 3.53→3.53 Å / SU ML: 1.4 / σ(F): 0.01 / Phase error: 64.36 / Stereochemistry target values: MLHL
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
|
 Movie
Movie Controller
Controller













 PDBj
PDBj