Entry Database : PDB / ID : 6i7nTitle Trypanothione Reductase from Leismania infantum in complex with TRL156 Trypanothione reductase Keywords / / / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species Leishmania infantum (eukaryote)Method / / / Resolution : 3.3 Å Authors Carriles, A.A. / Hermoso, J.A. Journal : Acs Infect Dis. / Year : 2019Title : Pyrrolopyrimidine vs Imidazole-Phenyl-Thiazole Scaffolds in Nonpeptidic Dimerization Inhibitors of Leishmania infantum Trypanothione Reductase.Authors : Revuelto, A. / Ruiz-Santaquiteria, M. / de Lucio, H. / Gamo, A. / Carriles, A.A. / Gutierrez, K.J. / Sanchez-Murcia, P.A. / Hermoso, J.A. / Gago, F. / Camarasa, M.J. / Jimenez-Ruiz, A. / Velazquez, S. History Deposition Nov 16, 2018 Deposition site / Processing site Revision 1.0 May 15, 2019 Provider / Type Revision 1.1 Jun 26, 2019 Group / Database references / Category / citation_author / pdbx_database_procItem _citation.journal_volume / _citation.page_first ... _citation.journal_volume / _citation.page_first / _citation.page_last / _citation_author.identifier_ORCID Revision 2.0 Oct 2, 2019 Group Advisory / Atomic model ... Advisory / Atomic model / Data collection / Database references / Derived calculations / Other / Polymer sequence / Source and taxonomy / Structure summary Category atom_site / atom_site_anisotrop ... atom_site / atom_site_anisotrop / cell / entity / entity_poly / entity_poly_seq / entity_src_gen / pdbx_entity_nonpoly / pdbx_nonpoly_scheme / pdbx_poly_seq_scheme / pdbx_struct_sheet_hbond / pdbx_unobs_or_zero_occ_residues / struct_asym / struct_conf / struct_conn / struct_mon_prot_cis / struct_ref / struct_ref_seq / struct_ref_seq_dif / struct_sheet_range / struct_site_gen Item _atom_site.label_entity_id / _atom_site.label_seq_id ... _atom_site.label_entity_id / _atom_site.label_seq_id / _atom_site_anisotrop.pdbx_label_seq_id / _cell.Z_PDB / _pdbx_entity_nonpoly.entity_id / _pdbx_nonpoly_scheme.entity_id / _pdbx_struct_sheet_hbond.range_1_label_seq_id / _pdbx_struct_sheet_hbond.range_2_label_seq_id / _struct_asym.entity_id / _struct_conf.beg_label_seq_id / _struct_conf.end_label_seq_id / _struct_conn.ptnr1_label_seq_id / _struct_conn.ptnr2_label_seq_id / _struct_mon_prot_cis.label_seq_id / _struct_mon_prot_cis.pdbx_label_seq_id_2 / _struct_ref_seq.ref_id / _struct_ref_seq.seq_align_beg / _struct_ref_seq.seq_align_end / _struct_sheet_range.beg_label_seq_id / _struct_sheet_range.end_label_seq_id / _struct_site_gen.label_seq_id Revision 2.1 Jan 24, 2024 Group / Database references / Refinement descriptionCategory chem_comp_atom / chem_comp_bond ... chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model / struct_ncs_dom_lim Item _database_2.pdbx_DOI / _database_2.pdbx_database_accession ... _database_2.pdbx_DOI / _database_2.pdbx_database_accession / _struct_ncs_dom_lim.beg_auth_comp_id / _struct_ncs_dom_lim.beg_label_asym_id / _struct_ncs_dom_lim.beg_label_comp_id / _struct_ncs_dom_lim.beg_label_seq_id / _struct_ncs_dom_lim.end_auth_comp_id / _struct_ncs_dom_lim.end_label_asym_id / _struct_ncs_dom_lim.end_label_comp_id / _struct_ncs_dom_lim.end_label_seq_id Revision 2.2 Oct 23, 2024 Group / Category / pdbx_modification_feature
Show all Show less
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information Leishmania infantum (eukaryote)
Leishmania infantum (eukaryote) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 3.3 Å
MOLECULAR REPLACEMENT / Resolution: 3.3 Å  Authors
Authors Citation
Citation Journal: Acs Infect Dis. / Year: 2019
Journal: Acs Infect Dis. / Year: 2019 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 6i7n.cif.gz
6i7n.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb6i7n.ent.gz
pdb6i7n.ent.gz PDB format
PDB format 6i7n.json.gz
6i7n.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads 6i7n_validation.pdf.gz
6i7n_validation.pdf.gz wwPDB validaton report
wwPDB validaton report 6i7n_full_validation.pdf.gz
6i7n_full_validation.pdf.gz 6i7n_validation.xml.gz
6i7n_validation.xml.gz 6i7n_validation.cif.gz
6i7n_validation.cif.gz https://data.pdbj.org/pub/pdb/validation_reports/i7/6i7n
https://data.pdbj.org/pub/pdb/validation_reports/i7/6i7n ftp://data.pdbj.org/pub/pdb/validation_reports/i7/6i7n
ftp://data.pdbj.org/pub/pdb/validation_reports/i7/6i7n
 Links
Links Assembly
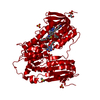
Assembly
 Components
Components Leishmania infantum (eukaryote) / Gene: TRYR, LINJ_05_0350 / Production host:
Leishmania infantum (eukaryote) / Gene: TRYR, LINJ_05_0350 / Production host: 



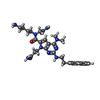





 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  ALBA
ALBA  / Beamline: XALOC / Wavelength: 0.97926 Å
/ Beamline: XALOC / Wavelength: 0.97926 Å Processing
Processing MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller


















 PDBj
PDBj





