Entry Database : PDB / ID : 5x94Title Crystal structure of SHP2_SH2-CagA EPIYA_D peptide complex Cag pathogenicity island protein Tyrosine-protein phosphatase non-receptor type 11 Keywords / / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species Homo sapiens (human)Helicobacter pylori (bacteria)Method / / / Resolution : 2.6 Å Authors Senda, M. / Senda, T. Journal : Cell Rep / Year : 2017Title : Differential Mechanisms for SHP2 Binding and Activation Are Exploited by Geographically Distinct Helicobacter pylori CagA Oncoproteins.Authors : Hayashi, T. / Senda, M. / Suzuki, N. / Nishikawa, H. / Ben, C. / Tang, C. / Nagase, L. / Inoue, K. / Senda, T. / Hatakeyama, M. History Deposition Mar 5, 2017 Deposition site / Processing site Revision 1.0 Sep 13, 2017 Provider / Type Revision 1.1 Oct 18, 2017 Group / Category / citation_authorItem _citation.country / _citation.journal_abbrev ... _citation.country / _citation.journal_abbrev / _citation.journal_id_CSD / _citation.journal_id_ISSN / _citation.journal_volume / _citation.page_first / _citation.page_last / _citation.pdbx_database_id_DOI / _citation.pdbx_database_id_PubMed / _citation.title / _citation.year Revision 1.2 Nov 22, 2023 Group / Database references / Refinement descriptionCategory chem_comp_atom / chem_comp_bond ... chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model Item / _database_2.pdbx_database_accessionRevision 1.3 Oct 23, 2024 Group / Category / pdbx_modification_feature
Show all Show less
 Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information Homo sapiens (human)
Homo sapiens (human)
 X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.6 Å
MOLECULAR REPLACEMENT / Resolution: 2.6 Å  Authors
Authors Citation
Citation Journal: Cell Rep / Year: 2017
Journal: Cell Rep / Year: 2017 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 5x94.cif.gz
5x94.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb5x94.ent.gz
pdb5x94.ent.gz PDB format
PDB format 5x94.json.gz
5x94.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/x9/5x94
https://data.pdbj.org/pub/pdb/validation_reports/x9/5x94 ftp://data.pdbj.org/pub/pdb/validation_reports/x9/5x94
ftp://data.pdbj.org/pub/pdb/validation_reports/x9/5x94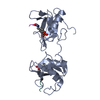

 Links
Links Assembly
Assembly


 Components
Components Homo sapiens (human) / Gene: PTPN11, PTP2C, SHPTP2
Homo sapiens (human) / Gene: PTPN11, PTP2C, SHPTP2

 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  Photon Factory
Photon Factory  / Beamline: BL-5A / Wavelength: 0.979 Å
/ Beamline: BL-5A / Wavelength: 0.979 Å Processing
Processing MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller













 PDBj
PDBj































