+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2qqg | ||||||
|---|---|---|---|---|---|---|---|
| Title | Hst2 bound to ADP-HPD, acetyllated histone H4 and nicotinamide | ||||||
 Components Components |
| ||||||
 Keywords Keywords | HYDROLASE / Hst2 / Sir2 / histone deacetylase / Metal-binding / NAD / Nucleus / Repressor / Transcription / Transcription regulation / Zinc | ||||||
| Function / homology |  Function and homology information Function and homology information: / negative regulation of mitotic recombination / HATs acetylate histones / RNA polymerase I upstream activating factor complex / histone H4K16 deacetylase activity, NAD-dependent / Condensation of Prophase Chromosomes / : / : / Assembly of the ORC complex at the origin of replication / HDACs deacetylate histones ...: / negative regulation of mitotic recombination / HATs acetylate histones / RNA polymerase I upstream activating factor complex / histone H4K16 deacetylase activity, NAD-dependent / Condensation of Prophase Chromosomes / : / : / Assembly of the ORC complex at the origin of replication / HDACs deacetylate histones / protein acetyllysine N-acetyltransferase / rDNA heterochromatin formation / histone deacetylase activity, NAD-dependent / Recruitment and ATM-mediated phosphorylation of repair and signaling proteins at DNA double strand breaks / Oxidative Stress Induced Senescence / RMTs methylate histone arginines / SUMOylation of chromatin organization proteins / RNA Polymerase I Promoter Escape / positive regulation of transcription by RNA polymerase I / nucleolar large rRNA transcription by RNA polymerase I / Estrogen-dependent gene expression / NAD+ binding / structural constituent of chromatin / nucleosome / nucleosome assembly / chromatin organization / protein heterodimerization activity / regulation of DNA-templated transcription / DNA binding / metal ion binding / identical protein binding / nucleus / cytoplasm Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.05 Å MOLECULAR REPLACEMENT / Resolution: 2.05 Å | ||||||
 Authors Authors | Marmorstein, R. / Sanders, B. / Zhao, K. / Slama, J. | ||||||
 Citation Citation |  Journal: Mol.Cell / Year: 2007 Journal: Mol.Cell / Year: 2007Title: Structural basis for nicotinamide inhibition and base exchange in sir2 enzymes. Authors: Sanders, B.D. / Zhao, K. / Slama, J.T. / Marmorstein, R. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2qqg.cif.gz 2qqg.cif.gz | 79.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2qqg.ent.gz pdb2qqg.ent.gz | 57.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2qqg.json.gz 2qqg.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/qq/2qqg https://data.pdbj.org/pub/pdb/validation_reports/qq/2qqg ftp://data.pdbj.org/pub/pdb/validation_reports/qq/2qqg ftp://data.pdbj.org/pub/pdb/validation_reports/qq/2qqg | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  2od2C  2od7C  2od9C  2qqfC  1szdS C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
|
- Components
Components
-Protein / Protein/peptide , 2 types, 2 molecules AB
| #1: Protein | Mass: 34794.828 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Gene: HST2 / Plasmid: pRSET-A / Production host:  References: UniProt: P53686, Hydrolases; Acting on carbon-nitrogen bonds, other than peptide bonds; In linear amides |
|---|---|
| #2: Protein/peptide | Mass: 1310.615 Da / Num. of mol.: 1 / Fragment: sequence database residues 13-23 / Source method: obtained synthetically Details: The H4 peptide was chemically synthesized using standard solid phase synthesis chemistry. The peptide is naturally found in Saccharomyces cerevisiae. References: UniProt: P02309 |
-Non-polymers , 4 types, 119 molecules 




| #3: Chemical | ChemComp-ZN / |
|---|---|
| #4: Chemical | ChemComp-A1R / |
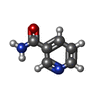
| #5: Chemical | ChemComp-NCA / |
| #6: Water | ChemComp-HOH / |
-Details
| Has protein modification | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.12 Å3/Da / Density % sol: 60.64 % |
|---|---|
| Crystal grow | Temperature: 298 K / pH: 5.6 Details: 2.0 M Ammonium Sulfate, 100 mM Na citrate, pH 5.6, 200 mM K/Na tartrate, VAPOR DIFFUSION, HANGING DROP, temperature 298K, pH 5.60 |
-Data collection
| Diffraction | Mean temperature: 93 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  CHESS CHESS  / Beamline: A1 / Wavelength: 0.9777 / Beamline: A1 / Wavelength: 0.9777 |
| Detector | Type: ADSC QUANTUM 210 / Detector: CCD / Date: Jul 16, 2006 |
| Radiation | Monochromator: SI(111) / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.9777 Å / Relative weight: 1 |
| Reflection | Resolution: 2.05→30 Å / Num. obs: 27580 / % possible obs: 99.9 % / Observed criterion σ(I): 6.1 / Redundancy: 6.1 % / Rmerge(I) obs: 0.111 / Rsym value: 0.117 / Net I/σ(I): 13.4 |
| Reflection shell | Resolution: 2.05→2.12 Å / Redundancy: 5.8 % / Rmerge(I) obs: 0.32 / % possible all: 99.6 |
-Phasing
| Phasing MR | Rfactor: 0.389 / Cor.coef. Fo:Fc: 0.658 / Cor.coef. Io to Ic: 0.404
|
|---|
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 1SZD Resolution: 2.05→28.34 Å / σ(F): 0
| ||||||||||||||||||||||||||||
| Solvent computation | Bsol: 59.07 Å2 | ||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 36.86 Å2
| ||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.05→28.34 Å
| ||||||||||||||||||||||||||||
| Xplor file |
|
 Movie
Movie Controller
Controller













 PDBj
PDBj











