[English] 日本語
 Yorodumi
Yorodumi- PDB-2cmr: Crystal structure of the HIV-1 neutralizing antibody D5 Fab bound... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2cmr | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of the HIV-1 neutralizing antibody D5 Fab bound to the gp41 inner-core mimetic 5-helix | ||||||
 Components Components |
| ||||||
 Keywords Keywords | IMMUNE SYSTEM / IMMUNOGLOBULIN COMPLEX / NEUTRALIZATION / IMMUNOGLOBULIN / ENVELOPE PROTEIN / HIV / GP41 / AIDS / MHC I / MEMBRANE / TRANSMEMBRANE / IMMUNOGLOBULIN DOMAIN | ||||||
| Function / homology |  Function and homology information Function and homology informationSynthesis and processing of ENV and VPU / symbiont-mediated evasion of host immune response / positive regulation of establishment of T cell polarity / Alpha-defensins / Dectin-2 family / Binding and entry of HIV virion / positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / actin filament organization / host cell endosome membrane ...Synthesis and processing of ENV and VPU / symbiont-mediated evasion of host immune response / positive regulation of establishment of T cell polarity / Alpha-defensins / Dectin-2 family / Binding and entry of HIV virion / positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / actin filament organization / host cell endosome membrane / Assembly Of The HIV Virion / Budding and maturation of HIV virion / clathrin-dependent endocytosis of virus by host cell / viral protein processing / receptor ligand activity / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / virion attachment to host cell / host cell plasma membrane / virion membrane / structural molecule activity / membrane Similarity search - Function | ||||||
| Biological species |   HUMAN IMMUNODEFICIENCY VIRUS 1 HUMAN IMMUNODEFICIENCY VIRUS 1 HOMO SAPIENS (human) HOMO SAPIENS (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2 Å MOLECULAR REPLACEMENT / Resolution: 2 Å | ||||||
 Authors Authors | Luftig, M.A. / Mattu, M. / Di Giovine, P. / Geleziunas, R. / Hrin, R. / Barbato, G. / Bianchi, E. / Miller, M.D. / Pessi, A. / Carfi, A. | ||||||
 Citation Citation |  Journal: Nat.Struct.Mol.Biol. / Year: 2006 Journal: Nat.Struct.Mol.Biol. / Year: 2006Title: Structural Basis for HIV-1 Neutralization by a Gp41 Fusion Intermediate-Directed Antibody Authors: Luftig, M.A. / Mattu, M. / Di Giovine, P. / Geleziunas, R. / Hrin, R. / Barbato, G. / Bianchi, E. / Miller, M.D. / Pessi, A. / Carfi, A. | ||||||
| History |
| ||||||
| Remark 700 | SHEET THE SHEET STRUCTURE OF THIS MOLECULE IS BIFURCATED. IN ORDER TO REPRESENT THIS FEATURE IN ... SHEET THE SHEET STRUCTURE OF THIS MOLECULE IS BIFURCATED. IN ORDER TO REPRESENT THIS FEATURE IN THE SHEET RECORDS BELOW, TWO SHEETS ARE DEFINED. |
- Structure visualization
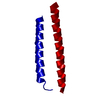
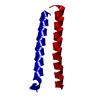
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2cmr.cif.gz 2cmr.cif.gz | 137.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2cmr.ent.gz pdb2cmr.ent.gz | 105.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2cmr.json.gz 2cmr.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  2cmr_validation.pdf.gz 2cmr_validation.pdf.gz | 446.8 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  2cmr_full_validation.pdf.gz 2cmr_full_validation.pdf.gz | 459.1 KB | Display | |
| Data in XML |  2cmr_validation.xml.gz 2cmr_validation.xml.gz | 26 KB | Display | |
| Data in CIF |  2cmr_validation.cif.gz 2cmr_validation.cif.gz | 37.9 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/cm/2cmr https://data.pdbj.org/pub/pdb/validation_reports/cm/2cmr ftp://data.pdbj.org/pub/pdb/validation_reports/cm/2cmr ftp://data.pdbj.org/pub/pdb/validation_reports/cm/2cmr | HTTPS FTP |
-Related structure data
| Related structure data |  1aikS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 25479.453 Da / Num. of mol.: 1 / Fragment: 5-HELIX, RESIDUES 543-582 AND 625-662 Source method: isolated from a genetically manipulated source Details: SEQUENCE DERIVED FROM GENBANK P04578 WHERE THREE REGIONS EACH EQUIVALENT TO AA 543-582 ARE INTERSPERSED BY TWO DIFFERENT REGIONS (EACH EQUIVALENT TO AA 625-662). LINKERS GGSGG AND GSSGG JOIN ...Details: SEQUENCE DERIVED FROM GENBANK P04578 WHERE THREE REGIONS EACH EQUIVALENT TO AA 543-582 ARE INTERSPERSED BY TWO DIFFERENT REGIONS (EACH EQUIVALENT TO AA 625-662). LINKERS GGSGG AND GSSGG JOIN THE REGIONS AND ALTERNATE STARTING WITH GGSGG. A METHIONINE INITIATES AND A TAIL CONTAINS GGHHHHHHG Source: (gene. exp.)   HUMAN IMMUNODEFICIENCY VIRUS 1 / Strain: HXB2 / Production host: HUMAN IMMUNODEFICIENCY VIRUS 1 / Strain: HXB2 / Production host:  |
|---|---|
| #2: Antibody | Mass: 22756.447 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  HOMO SAPIENS (human) / Description: IGG DERIVED FROM HUMAN B CELL LIBRARY / Plasmid: PEU3.2, PEU1.2 / Cell line (production host): 293-EBNA / Production host: HOMO SAPIENS (human) / Description: IGG DERIVED FROM HUMAN B CELL LIBRARY / Plasmid: PEU3.2, PEU1.2 / Cell line (production host): 293-EBNA / Production host:  Homo sapiens (human) Homo sapiens (human) |
| #3: Antibody | Mass: 22689.236 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  HOMO SAPIENS (human) / Description: IGG DERIVED FROM HUMAN B CELL LIBRARY / Plasmid: PEU3.2, PEU1.2 / Cell line (production host): 293-EBNA / Production host: HOMO SAPIENS (human) / Description: IGG DERIVED FROM HUMAN B CELL LIBRARY / Plasmid: PEU3.2, PEU1.2 / Cell line (production host): 293-EBNA / Production host:  Homo sapiens (human) Homo sapiens (human) |
| #4: Chemical | ChemComp-GOL / |
| #5: Water | ChemComp-HOH / |
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.5 Å3/Da / Density % sol: 50 % |
|---|---|
| Crystal grow | pH: 8.5 Details: 18% (W/V) PEG 4000, 0.2 M TRI-SODIUM CITRATE DIHYDRATE, 0.1 M TRIS-HCL / PH = 8.5 |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ESRF ESRF  / Beamline: ID14-2 / Wavelength: 0.933 / Beamline: ID14-2 / Wavelength: 0.933 |
| Detector | Type: MARRESEARCH / Detector: CCD / Date: Apr 21, 2004 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.933 Å / Relative weight: 1 |
| Reflection | Resolution: 2→33.5 Å / Num. obs: 41190 / % possible obs: 90 % / Observed criterion σ(I): 3.5 / Redundancy: 2.6 % / Rmerge(I) obs: 0.11 / Net I/σ(I): 8 |
| Reflection shell | Resolution: 2→2.1 Å / Redundancy: 2.1 % / Rmerge(I) obs: 0.51 / Mean I/σ(I) obs: 1.7 / % possible all: 90 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 1AIK Resolution: 2→33.52 Å / Cor.coef. Fo:Fc: 0.939 / Cor.coef. Fo:Fc free: 0.907 / SU B: 4.936 / SU ML: 0.135 / Cross valid method: THROUGHOUT / ESU R: 0.21 / ESU R Free: 0.186 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.4 Å / Solvent model: BABINET MODEL WITH MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 28.54 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2→33.52 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller






















 PDBj
PDBj












