+ Open data
Open data
- Basic information
Basic information
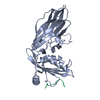
| Entry | Database: PDB / ID: 1utc | ||||||
|---|---|---|---|---|---|---|---|
| Title | Clathrin terminal domain complexed with TLPWDLWTT | ||||||
 Components Components |
| ||||||
 Keywords Keywords | ENDOCYTOSIS / CLATHRIN / CYTOSKELETON | ||||||
| Function / homology |  Function and homology information Function and homology informationRetrograde neurotrophin signalling / Recycling pathway of L1 / WNT5A-dependent internalization of FZD4 / WNT5A-dependent internalization of FZD2, FZD5 and ROR2 / LDL clearance / Gap junction degradation / Formation of annular gap junctions / RHOU GTPase cycle / RHOV GTPase cycle / Golgi Associated Vesicle Biogenesis ...Retrograde neurotrophin signalling / Recycling pathway of L1 / WNT5A-dependent internalization of FZD4 / WNT5A-dependent internalization of FZD2, FZD5 and ROR2 / LDL clearance / Gap junction degradation / Formation of annular gap junctions / RHOU GTPase cycle / RHOV GTPase cycle / Golgi Associated Vesicle Biogenesis / clathrin coat of trans-Golgi network vesicle / Lysosome Vesicle Biogenesis / clathrin light chain binding / negative regulation of hyaluronan biosynthetic process / clathrin complex / MHC class II antigen presentation / VLDLR internalisation and degradation / clathrin coat of coated pit / clathrin coat disassembly / Cargo recognition for clathrin-mediated endocytosis / membrane coat / clathrin-coated endocytic vesicle / clathrin coat assembly / leading edge membrane / Clathrin-mediated endocytosis / arrestin family protein binding / synaptic vesicle endocytosis / receptor-mediated endocytosis / intracellular protein transport / phospholipid binding / autophagy / spindle / endocytosis / disordered domain specific binding / synaptic vesicle / melanosome / mitotic cell cycle / synaptic vesicle membrane / actin cytoskeleton / Clathrin-mediated endocytosis / chemical synaptic transmission / protein domain specific binding / cell division / structural molecule activity / mitochondrion / identical protein binding / plasma membrane / cytosol Similarity search - Function | ||||||
| Biological species |   HOMO SAPIENS (human) HOMO SAPIENS (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.3 Å MOLECULAR REPLACEMENT / Resolution: 2.3 Å | ||||||
 Authors Authors | Miele, A.E. / Evans, P.R. / Owen, D.J. | ||||||
 Citation Citation |  Journal: Nat. Struct. Mol. Biol. / Year: 2004 Journal: Nat. Struct. Mol. Biol. / Year: 2004Title: Two distinct interaction motifs in amphiphysin bind two independent sites on the clathrin terminal domain beta-propeller. Authors: Miele, A.E. / Watson, P.J. / Evans, P.R. / Traub, L.M. / Owen, D.J. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1utc.cif.gz 1utc.cif.gz | 157 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1utc.ent.gz pdb1utc.ent.gz | 124.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1utc.json.gz 1utc.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ut/1utc https://data.pdbj.org/pub/pdb/validation_reports/ut/1utc ftp://data.pdbj.org/pub/pdb/validation_reports/ut/1utc ftp://data.pdbj.org/pub/pdb/validation_reports/ut/1utc | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1c9iS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
| ||||||||
| Noncrystallographic symmetry (NCS) | NCS oper: (Code: given Matrix: (-0.999979, -0.005338, 0.00362), Vector: Details | CLATHRIN HEAVY CHAIN IS A MONOMER BIOLOGICALLY BUTIS CLASSIFIED AS A DIMER IN THIS ENTRY SINCE EVERYMOLECULE OF CLATHRIN (CHAINS A AND B) ARE BOUND TOA PEPTIDE. | |
- Components
Components
| #1: Antibody | Mass: 40396.340 Da / Num. of mol.: 2 / Fragment: TERMINAL DOMAIN, RESIDUES 1-363 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   #2: Protein/peptide | Mass: 1132.264 Da / Num. of mol.: 2 / Fragment: W-BOX, RESIDUES 379-387 / Source method: obtained synthetically / Details: PEPTIDE FROM AMPHIPHYSIN 1 / Source: (synth.)  HOMO SAPIENS (human) / References: UniProt: P49418 HOMO SAPIENS (human) / References: UniProt: P49418#3: Water | ChemComp-HOH / | Compound details | CLATHRIN IS THE MAJOR PROTEIN OF THE POLYHEDRAL COAT OF COATED PITS & VESICLES. TWO DIFFERENT ...CLATHRIN IS THE MAJOR PROTEIN OF THE POLYHEDRAL | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.38 Å3/Da / Density % sol: 48.28 % | |||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Method: vapor diffusion, hanging drop / pH: 9 Details: VAPOUR DIFFUSION OVER A RESERVOIR CONTAINING 20% PEG8000, 200 MM MGCL2, 4 MM DTT,100 MM CAPS PH 9, DRIED UP DROP | |||||||||||||||||||||||||||||||||||
| Crystal grow | *PLUS pH: 9 / Method: vapor diffusion, hanging drop | |||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SRS SRS  / Beamline: PX9.6 / Wavelength: 0.87 / Beamline: PX9.6 / Wavelength: 0.87 |
| Detector | Type: ADSC CCD / Detector: CCD / Details: MIRRORS |
| Radiation | Monochromator: SI / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.87 Å / Relative weight: 1 |
| Reflection | Resolution: 2.3→24.92 Å / Num. obs: 37228 / % possible obs: 99.4 % / Redundancy: 5.84 % / Rmerge(I) obs: 0.057 / Net I/σ(I): 9.3131 |
| Reflection shell | Resolution: 2.3→2.42 Å / Redundancy: 4.33 % / Rmerge(I) obs: 0.33 / Mean I/σ(I) obs: 2.02 / % possible all: 97.4 |
| Reflection | *PLUS Highest resolution: 2.3 Å / Lowest resolution: 29.9 Å / Num. obs: 37078 / Redundancy: 5.8 % / Num. measured all: 495661 / Rmerge(I) obs: 0.076 |
| Reflection shell | *PLUS % possible obs: 97.2 % / Rmerge(I) obs: 0.329 / Mean I/σ(I) obs: 5.3 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 1C9I Resolution: 2.3→105.41 Å / Cor.coef. Fo:Fc: 0.934 / Cor.coef. Fo:Fc free: 0.892 / SU B: 7.03 / SU ML: 0.173 / Cross valid method: THROUGHOUT / ESU R: 0.334 / ESU R Free: 0.245 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å / Solvent model: BABINET MODEL WITH MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 21.21 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.3→105.41 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller














 PDBj
PDBj


