[English] 日本語
 Yorodumi
Yorodumi- PDB-1qop: CRYSTAL STRUCTURE OF WILD-TYPE TRYPTOPHAN SYNTHASE COMPLEXED WITH... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1qop | ||||||
|---|---|---|---|---|---|---|---|
| Title | CRYSTAL STRUCTURE OF WILD-TYPE TRYPTOPHAN SYNTHASE COMPLEXED WITH INDOLE PROPANOL PHOSPHATE | ||||||
 Components Components | (TRYPTOPHAN SYNTHASE ...) x 2 | ||||||
 Keywords Keywords | LYASE / CARBON-OXYGEN LYASE / TRYPTOPHAN BIOSYNTHESIS / PYRIDOXAL PHOSPHATE | ||||||
| Function / homology |  Function and homology information Function and homology informationtryptophan synthase / tryptophan synthase activity / L-tryptophan biosynthetic process / identical protein binding / cytosol / cytoplasm Similarity search - Function | ||||||
| Biological species |  SALMONELLA TYPHIMURIUM (bacteria) SALMONELLA TYPHIMURIUM (bacteria) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.4 Å MOLECULAR REPLACEMENT / Resolution: 1.4 Å | ||||||
 Authors Authors | Weyand, M. / Schlichting, I. | ||||||
 Citation Citation |  Journal: Biochemistry / Year: 1999 Journal: Biochemistry / Year: 1999Title: Crystal Structure of Wild-Type Tryptophan Synthase Complexed with the Natural Substrate Indole-3-Glycerol Phosphate Authors: Weyand, M. / Schlichting, I. | ||||||
| History |
| ||||||
| Remark 650 | HELIX DETERMINATION METHOD: DSSP, KABSCH & SANDER | ||||||
| Remark 700 | SHEET DETERMINATION METHOD: DSSP, KABSCH & SANDER |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1qop.cif.gz 1qop.cif.gz | 297.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1qop.ent.gz pdb1qop.ent.gz | 237.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1qop.json.gz 1qop.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/qo/1qop https://data.pdbj.org/pub/pdb/validation_reports/qo/1qop ftp://data.pdbj.org/pub/pdb/validation_reports/qo/1qop ftp://data.pdbj.org/pub/pdb/validation_reports/qo/1qop | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1qoqC  2wsyS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| Unit cell |
| ||||||||
| Details | BIOMOLECULE THE BIOLOGICALLY ACTIVE MOLECULE IS ATETRAMER OF TWO ALPHA AND TWO BETA CHAINS. |
- Components
Components
-TRYPTOPHAN SYNTHASE ... , 2 types, 2 molecules AB
| #1: Protein | Mass: 28698.797 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Details: INHIBITOR INDOLE PROPANOL PHOSPHATE BOUND TO THE ALPHA SITE Source: (gene. exp.)  SALMONELLA TYPHIMURIUM (bacteria) / Gene: TRPA / Plasmid: PSTB7 / Production host: SALMONELLA TYPHIMURIUM (bacteria) / Gene: TRPA / Plasmid: PSTB7 / Production host:  |
|---|---|
| #2: Protein | Mass: 42787.688 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  SALMONELLA TYPHIMURIUM (bacteria) / Gene: TRPB / Plasmid: PSTB7 / Production host: SALMONELLA TYPHIMURIUM (bacteria) / Gene: TRPB / Plasmid: PSTB7 / Production host:  References: UniProt: P00933, UniProt: P0A2K1*PLUS, tryptophan synthase |
-Non-polymers , 4 types, 804 molecules 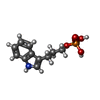






| #3: Chemical | ChemComp-IPL / |
|---|---|
| #4: Chemical | ChemComp-PLP / |
| #5: Chemical | ChemComp-NA / |
| #6: Water | ChemComp-HOH / |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.58 Å3/Da / Density % sol: 52.4 % | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Method: vapor diffusion, hanging drop / pH: 7.8 Details: ENZYME SOLUTION: 10 MG/ML TRPS IN 50 MM BICINE PH 7.8, 1 MM EDTA, 5 MM DITHIOERYTHRITOL, 20 MUM PYRIDOXAL-5'-PHOSPHATE. RESERVOIR SOLUTION: 50 MM BICINE PH 7.8, 5 MM EDTA, 5 MM ...Details: ENZYME SOLUTION: 10 MG/ML TRPS IN 50 MM BICINE PH 7.8, 1 MM EDTA, 5 MM DITHIOERYTHRITOL, 20 MUM PYRIDOXAL-5'-PHOSPHATE. RESERVOIR SOLUTION: 50 MM BICINE PH 7.8, 5 MM EDTA, 5 MM DITHIOERYTHRITOL, 0.1 MM PYRIDOXAL-5'-PHOSPHATE, 2 MM SPERMINE, 8-12 % PEG 8000. HANGING DROP GEOMETRY, CRYSTALLIZATION DROP CONSISTED AN INITIAL INDOLE PROPANOLE PHOSPHATE (IPL) CONCENTRATION OF 7 MM | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Method: vapor diffusion, hanging drop | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  EMBL/DESY, HAMBURG EMBL/DESY, HAMBURG  / Beamline: X11 / Wavelength: 0.9076 / Beamline: X11 / Wavelength: 0.9076 |
| Detector | Type: MARRESEARCH / Detector: IMAGE PLATE / Date: Jun 15, 1998 / Details: SYNCHROTRON |
| Radiation | Monochromator: SYNCHROTRON / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.9076 Å / Relative weight: 1 |
| Reflection | Resolution: 1.4→30.3 Å / Num. obs: 133934 / % possible obs: 93.2 % / Observed criterion σ(I): -3 / Redundancy: 2.2 % / Rsym value: 0.057 / Net I/σ(I): 10.25 |
| Reflection shell | Resolution: 1.4→1.5 Å / Redundancy: 1.89 % / Mean I/σ(I) obs: 2.36 / Rsym value: 0.258 / % possible all: 80.8 |
| Reflection | *PLUS Num. measured all: 294137 / Rmerge(I) obs: 0.057 |
| Reflection shell | *PLUS % possible obs: 80.8 % / Rmerge(I) obs: 0.258 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 2WSY Resolution: 1.4→20 Å / SU B: 0.91781 / SU ML: 0.03585 / Cross valid method: THROUGHOUT / σ(F): 0 / ESU R: 0.06129 / ESU R Free: 0.05539 Details: RESIDUES A156, A157 AND A158 WERE MODELED IN TWO CONFORMATIONS INCLUDING THE MAIN CHAIN ATOMS. THE SIDE CHAINS OF RESIDUES ILE A30, GLU A31, CYS A154, SER A178 AND GLU A254 WERE MODELED IN ...Details: RESIDUES A156, A157 AND A158 WERE MODELED IN TWO CONFORMATIONS INCLUDING THE MAIN CHAIN ATOMS. THE SIDE CHAINS OF RESIDUES ILE A30, GLU A31, CYS A154, SER A178 AND GLU A254 WERE MODELED IN TWO CONFORMATIONS THE SIDE CHAINS OF RESIDUES GLU B11, MET B22, LYS B37, LYS B50, LYS B61, ILE B65, MET B152, THR B165, LYS B219, MET B240, HIS B260, ARG B341, GLU B367 AND ARG B379 WERE MODELED IN TWO CONFORMATIONS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 18 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.4→20 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name: REFMAC / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS Rfactor obs: 0.15 / Rfactor Rwork: 0.15 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS |
 Movie
Movie Controller
Controller

































 PDBj
PDBj



