+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1odv | ||||||
|---|---|---|---|---|---|---|---|
| Title | Photoactive yellow protein 1-25 deletion mutant | ||||||
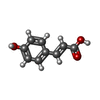
 Components Components | PHOTOACTIVE YELLOW PROTEIN | ||||||
 Keywords Keywords | SIGNALLING / PHOTOACTIVITY / P-COUMARIC ACID | ||||||
| Function / homology |  Function and homology information Function and homology informationphotoreceptor activity / phototransduction / regulation of DNA-templated transcription / identical protein binding Similarity search - Function | ||||||
| Biological species |  ECTOTHIORHODOSPIRA HALOPHILA (bacteria) ECTOTHIORHODOSPIRA HALOPHILA (bacteria) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.14 Å MOLECULAR REPLACEMENT / Resolution: 1.14 Å | ||||||
 Authors Authors | Vreede, J. / Van Der horst, M.A. / Hellingwerf, K.J. / Crielaard, W. / Van Aalten, D.M.F. | ||||||
 Citation Citation |  Journal: J.Biol.Chem. / Year: 2003 Journal: J.Biol.Chem. / Year: 2003Title: Pas Domains.Common Structure and Common Flexibility Authors: Vreede, J. / Van Der Horst, M.A. / Hellingwerf, K.J. / Crielaard, W. / Van Aalten, D.M.F. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1odv.cif.gz 1odv.cif.gz | 111.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1odv.ent.gz pdb1odv.ent.gz | 85.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1odv.json.gz 1odv.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/od/1odv https://data.pdbj.org/pub/pdb/validation_reports/od/1odv ftp://data.pdbj.org/pub/pdb/validation_reports/od/1odv ftp://data.pdbj.org/pub/pdb/validation_reports/od/1odv | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  2phyS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| |||||||||
| 2 | 
| |||||||||
| Unit cell |
| |||||||||
| Components on special symmetry positions |
|
- Components
Components
| #1: Protein | Mass: 11181.659 Da / Num. of mol.: 2 / Fragment: RESIDUES 26-125 Source method: isolated from a genetically manipulated source Details: P-COUMARIC ACID Source: (gene. exp.)  ECTOTHIORHODOSPIRA HALOPHILA (bacteria) ECTOTHIORHODOSPIRA HALOPHILA (bacteria)Production host:  #2: Chemical | #3: Water | ChemComp-HOH / | Compound details | PHOTOACTIVE BLUE-LIGHT PROTEIN THAT PROBABLY FUNCTIONS AS A PHOTORECEPTOR FOR A NEGATIVE PHOTOTAXIS ...PHOTOACTIV | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.07 Å3/Da / Density % sol: 40.6 % | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | pH: 7.5 Details: CRYSTALS WERE GROWN BY EQUILIBRATION OF 1 ML OF 30 MG/ML PROTEIN WITH 1 ML OF MOTHER LIQUOR (1.8 M AMMONIUM SULPHATE, 10 MM COCL[2], 100 MM MES PH 6.5) AGAINST A 1 ML RESERVOIR OF MOTHER ...Details: CRYSTALS WERE GROWN BY EQUILIBRATION OF 1 ML OF 30 MG/ML PROTEIN WITH 1 ML OF MOTHER LIQUOR (1.8 M AMMONIUM SULPHATE, 10 MM COCL[2], 100 MM MES PH 6.5) AGAINST A 1 ML RESERVOIR OF MOTHER LIQUOR. CRYSTALS APPEARED AFTER 2-3 DAYS A LARGEST DIMENSION OF 0.4 MM. | ||||||||||||||||||||||||||||||
| Crystal grow | *PLUS pH: 6.5 / Method: unknown | ||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ESRF ESRF  / Beamline: ID14-1 / Wavelength: 0.934 / Beamline: ID14-1 / Wavelength: 0.934 |
| Detector | Date: Nov 15, 2000 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.934 Å / Relative weight: 1 |
| Reflection | Resolution: 1.14→17 Å / Num. obs: 299780 / % possible obs: 94.4 % / Redundancy: 4 % / Rmerge(I) obs: 0.06 / Net I/σ(I): 13.5 |
| Reflection shell | Resolution: 1.14→1.18 Å / Redundancy: 3.3 % / Rmerge(I) obs: 0.217 / Mean I/σ(I) obs: 4.3 / % possible all: 89.2 |
| Reflection | *PLUS Num. obs: 75208 / Num. measured all: 299780 |
| Reflection shell | *PLUS % possible obs: 89.2 % |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 2PHY Resolution: 1.14→17 Å / Num. parameters: 17869 / Num. restraintsaints: 21180 / Cross valid method: FREE R-VALUE / σ(F): 0 / Stereochemistry target values: ENGH AND HUBER
| |||||||||||||||||||||||||||||||||
| Solvent computation | Solvent model: MOEWS & KRETSINGER, J.MOL.BIOL.91(1973)201-2 | |||||||||||||||||||||||||||||||||
| Refine analyze | Num. disordered residues: 7 / Occupancy sum hydrogen: 1459 / Occupancy sum non hydrogen: 1919 | |||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.14→17 Å
| |||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||
| Software | *PLUS Name: SHELXL-97 / Classification: refinement | |||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
|
 Movie
Movie Controller
Controller























 PDBj
PDBj