[English] 日本語
 Yorodumi
Yorodumi- PDB-1nr2: High resolution crystal structures of thymus and activation-regul... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1nr2 | ||||||
|---|---|---|---|---|---|---|---|
| Title | High resolution crystal structures of thymus and activation-regulated chemokine | ||||||
 Components Components | Thymus and activation-regulated chemokine | ||||||
 Keywords Keywords | CYTOKINE / TARC / chemokine / CC-chemokine / chemotaxis | ||||||
| Function / homology |  Function and homology information Function and homology informationchemokine activity / Chemokine receptors bind chemokines / chemotaxis / cell-cell signaling / immune response / inflammatory response / signaling receptor binding / extracellular space / extracellular region Similarity search - Function | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.18 Å MOLECULAR REPLACEMENT / Resolution: 2.18 Å | ||||||
 Authors Authors | Asojo, O.A. / Boulegue, C. / Hoover, D.M. / Lu, W. / Lubkowski, J. | ||||||
 Citation Citation |  Journal: Acta Crystallogr.,Sect.D / Year: 2003 Journal: Acta Crystallogr.,Sect.D / Year: 2003Title: Structures of thymus and activation-regulated chemokine (TARC). Authors: Asojo, O.A. / Boulegue, C. / Hoover, D.M. / Lu, W. / Lubkowski, J. #1:  Journal: Acta Crystallogr.,Sect.D / Year: 2003 Journal: Acta Crystallogr.,Sect.D / Year: 2003Title: Crystallization and preliminary X-ray studies of thymus and activation-regulated chemokine Authors: Asojo, O.A. / Hoover, D. / Boulegue, C. / Cater, S. / Lu, W. / Lubkowski, J. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1nr2.cif.gz 1nr2.cif.gz | 42 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1nr2.ent.gz pdb1nr2.ent.gz | 29.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1nr2.json.gz 1nr2.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/nr/1nr2 https://data.pdbj.org/pub/pdb/validation_reports/nr/1nr2 ftp://data.pdbj.org/pub/pdb/validation_reports/nr/1nr2 ftp://data.pdbj.org/pub/pdb/validation_reports/nr/1nr2 | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 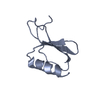
| ||||||||||
| 2 | 
| ||||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 8096.292 Da / Num. of mol.: 2 / Source method: obtained synthetically / Details: This sequence occurs naturally in humans / References: UniProt: Q92583 #2: Water | ChemComp-HOH / | Has protein modification | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 1.93 Å3/Da / Density % sol: 35.6 % | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Temperature: 288 K / Method: vapor diffusion, hanging drop / pH: 5.6 Details: 0.17M ammonium acetate, 0.085M trisodium citrate, 25.5% PEG 4000, 15% w/v glycerol, pH 5.6, VAPOR DIFFUSION, HANGING DROP, temperature 288K | ||||||||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Temperature: 285 K / Method: vapor diffusion, hanging drop | ||||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 90 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  NSLS NSLS  / Beamline: X9B / Wavelength: 0.979 Å / Beamline: X9B / Wavelength: 0.979 Å |
| Detector | Type: ADSC QUANTUM 4 / Detector: CCD / Date: Jun 4, 2002 |
| Radiation | Monochromator: Mirrors / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.979 Å / Relative weight: 1 |
| Reflection | Resolution: 2.18→10 Å / Num. all: 6647 / Num. obs: 6647 / % possible obs: 92.2 % / Observed criterion σ(F): 0 / Observed criterion σ(I): 0 / Redundancy: 3.5 % / Rmerge(I) obs: 0.062 / Rsym value: 0.064 / Net I/σ(I): 19 |
| Reflection shell | Resolution: 2.18→2.26 Å / Redundancy: 2.5 % / Rmerge(I) obs: 0.194 / Rsym value: 0.271 / % possible all: 93.4 |
| Reflection | *PLUS Lowest resolution: 25 Å / % possible obs: 93.5 % / Redundancy: 5.5 % |
| Reflection shell | *PLUS % possible obs: 92.2 % / Mean I/σ(I) obs: 2.1 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: RANTES Resolution: 2.18→10 Å / Num. parameters: 4927 / Num. restraintsaints: 4188 / Cross valid method: FREE R / σ(F): 0 / Stereochemistry target values: ENGH AND HUBER
| |||||||||||||||||||||||||||||||||
| Refine analyze | Num. disordered residues: 0 / Occupancy sum hydrogen: 0 / Occupancy sum non hydrogen: 1231 | |||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.18→10 Å
| |||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||
| Software | *PLUS Name: SHELXL / Version: 97 / Classification: refinement | |||||||||||||||||||||||||||||||||
| Refinement | *PLUS Lowest resolution: 10 Å / Rfactor Rfree: 0.277 / Rfactor Rwork: 0.198 | |||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | |||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS | |||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
| |||||||||||||||||||||||||||||||||
| LS refinement shell | *PLUS Highest resolution: 2.18 Å / Lowest resolution: 2.26 Å / Rfactor Rfree: 0.391 / Rfactor Rwork: 0.26 |
 Movie
Movie Controller
Controller











 PDBj
PDBj

