[English] 日本語
 Yorodumi
Yorodumi- PDB-1nr4: High resolution crystal structures of thymus and activation-regul... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1nr4 | ||||||
|---|---|---|---|---|---|---|---|
| Title | High resolution crystal structures of thymus and activation-regulated chemokine | ||||||
 Components Components | Thymus and activation-regulated chemokine | ||||||
 Keywords Keywords | CYTOKINE / TARC / chemokine / CC-chemokine / chemotaxis | ||||||
| Function / homology |  Function and homology information Function and homology informationchemokine activity / Chemokine receptors bind chemokines / chemotaxis / cell-cell signaling / immune response / inflammatory response / signaling receptor binding / extracellular space / extracellular region Similarity search - Function | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 1.72 Å MOLECULAR REPLACEMENT / Resolution: 1.72 Å | ||||||
 Authors Authors | Asojo, O.A. / Boulegue, C. / Hoover, D.M. / Lu, W. / Lubkowski, J. | ||||||
 Citation Citation |  Journal: Acta Crystallogr.,Sect.D / Year: 2003 Journal: Acta Crystallogr.,Sect.D / Year: 2003Title: Structures of thymus and activation-regulated chemokine (TARC). Authors: Asojo, O.A. / Boulegue, C. / Hoover, D.M. / Lu, W. / Lubkowski, J. #1:  Journal: Acta Crystallogr.,Sect.D / Year: 2003 Journal: Acta Crystallogr.,Sect.D / Year: 2003Title: Crystallization and preliminary X-ray studies of thymus and activation-regulated chemokine Authors: Asojo, O.A. / Hoover, D. / Boulegue, C. / Cater, S. / Lu, W. / Lubkowski, J. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1nr4.cif.gz 1nr4.cif.gz | 129.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1nr4.ent.gz pdb1nr4.ent.gz | 103.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1nr4.json.gz 1nr4.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/nr/1nr4 https://data.pdbj.org/pub/pdb/validation_reports/nr/1nr4 ftp://data.pdbj.org/pub/pdb/validation_reports/nr/1nr4 ftp://data.pdbj.org/pub/pdb/validation_reports/nr/1nr4 | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||||
| 2 | 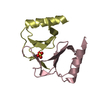
| ||||||||||
| 3 | 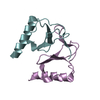
| ||||||||||
| 4 | 
| ||||||||||
| 5 | 
| ||||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 8096.292 Da / Num. of mol.: 8 / Source method: obtained synthetically / Details: This sequence occurs naturally in humans / References: UniProt: Q92583 #2: Chemical | ChemComp-SO4 / #3: Water | ChemComp-HOH / | Has protein modification | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.52 Å3/Da / Density % sol: 50.8 % | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Temperature: 285 K / Method: vapor diffusion, hanging drop / pH: 4.6 Details: 0.16M ammonium sulfate, 0.08M Sodium acetate, 20% PEG 4000, 15% glycerol, pH 4.6, VAPOR DIFFUSION, HANGING DROP, temperature 285K | ||||||||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Temperature: 285 K / pH: 5.6 / Method: vapor diffusion, hanging drop | ||||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 90 K |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU RU200 / Wavelength: 1.5418 Å ROTATING ANODE / Type: RIGAKU RU200 / Wavelength: 1.5418 Å |
| Detector | Type: MARRESEARCH / Detector: IMAGE PLATE / Date: Aug 1, 2002 / Details: Mirrors |
| Radiation | Monochromator: Mirrors / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.5418 Å / Relative weight: 1 |
| Reflection | Resolution: 1.72→23 Å / Num. all: 70685 / Num. obs: 65586 / % possible obs: 92.8 % / Observed criterion σ(I): 0 / Redundancy: 7 % / Rmerge(I) obs: 0.063 / Rsym value: 0.053 / Net I/σ(I): 12 |
| Reflection shell | Resolution: 1.72→1.78 Å / Redundancy: 3 % / Rmerge(I) obs: 0.241 / Mean I/σ(I) obs: 5.1 / Num. unique all: 5781 / Rsym value: 0.26 / % possible all: 81.5 |
| Reflection | *PLUS Lowest resolution: 20 Å / % possible obs: 92.79 % / Rmerge(I) obs: 0.075 |
| Reflection shell | *PLUS Lowest resolution: 1.84 Å / % possible obs: 92.9 % / Rmerge(I) obs: 0.279 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: RANTES Resolution: 1.72→24.92 Å / Cor.coef. Fo:Fc: 0.957 / Cor.coef. Fo:Fc free: 0.937 / SU B: 2.349 / SU ML: 0.077 / Cross valid method: THROUGHOUT / σ(F): 0 / ESU R: 0.121 / ESU R Free: 0.12 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.4 Å / Solvent model: BABINET MODEL WITH MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 31.584 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.72→24.92 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 1.72→1.84 Å / Total num. of bins used: 8
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS Lowest resolution: 20 Å / % reflection Rfree: 5 % / Rfactor Rfree: 0.241 / Rfactor Rwork: 0.198 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | *PLUS Rfactor Rfree: 0.354 / Rfactor Rwork: 0.275 |
 Movie
Movie Controller
Controller











 PDBj
PDBj



