+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1cfv | ||||||
|---|---|---|---|---|---|---|---|
| Title | MONOCLONAL ANTIBODY FRAGMENT FV4155 FROM E. COLI | ||||||
 Components Components | (MONOCLONAL ANTIBODY FV4155) x 2 | ||||||
 Keywords Keywords | IMMUNOGLOBULIN / FV FRAGMENT / STEROID HORMONE / FINE SPECIFICITY | ||||||
| Function / homology |  Function and homology information Function and homology informationimmunoglobulin mediated immune response / immunoglobulin complex / antigen binding / adaptive immune response / immune response / extracellular region Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 2.1 Å MOLECULAR REPLACEMENT / Resolution: 2.1 Å | ||||||
 Authors Authors | Trinh, C.H. / Phillips, S.E.V. | ||||||
 Citation Citation |  Journal: Structure / Year: 1997 Journal: Structure / Year: 1997Title: Antibody fragment Fv4155 bound to two closely related steroid hormones: the structural basis of fine specificity. Authors: Trinh, C.H. / Hemmington, S.D. / Verhoeyen, M.E. / Phillips, S.E. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1cfv.cif.gz 1cfv.cif.gz | 65.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1cfv.ent.gz pdb1cfv.ent.gz | 46.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1cfv.json.gz 1cfv.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/cf/1cfv https://data.pdbj.org/pub/pdb/validation_reports/cf/1cfv ftp://data.pdbj.org/pub/pdb/validation_reports/cf/1cfv ftp://data.pdbj.org/pub/pdb/validation_reports/cf/1cfv | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1bfvC  2bfvC  1nbvS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||||||
| Unit cell |
| ||||||||||||
| Components on special symmetry positions |
|
- Components
Components
| #1: Antibody | Mass: 12437.998 Da / Num. of mol.: 1 / Fragment: FV FRAGMENT Source method: isolated from a genetically manipulated source Source: (gene. exp.)   | ||||||
|---|---|---|---|---|---|---|---|
| #2: Antibody | Mass: 13191.760 Da / Num. of mol.: 1 / Fragment: FV FRAGMENT Source method: isolated from a genetically manipulated source Source: (gene. exp.)   | ||||||
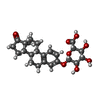
| #3: Chemical | | #4: Chemical | ChemComp-E3G / | #5: Water | ChemComp-HOH / | Has protein modification | Y | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.36 Å3/Da / Density % sol: 48 % | ||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | pH: 7 Details: PROTEIN WAS CRYSTALLIZED FROM 18% (W/V) PEG 8000, 200MM ZN ACETATE AND 100MM NA CACODYLATE, PH 7.0 | ||||||||||||||||||||||||||||||||||||||||
| Crystal | *PLUS Density % sol: 48 % | ||||||||||||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Method: vapor diffusion, hanging drop | ||||||||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU RUH2R / Wavelength: 1.5418 ROTATING ANODE / Type: RIGAKU RUH2R / Wavelength: 1.5418 |
| Detector | Type: RIGAKU RAXIS IIC / Detector: IMAGE PLATE / Date: Jan 1, 1996 / Details: MSC/YALE MIRRORS |
| Radiation | Monochromator: NI FILTER / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.5418 Å / Relative weight: 1 |
| Reflection | Resolution: 2.1→20 Å / Num. obs: 14295 / % possible obs: 97.1 % / Observed criterion σ(I): 3 / Redundancy: 7.4 % / Rsym value: 0.064 / Net I/σ(I): 9.5 |
| Reflection shell | Resolution: 2.1→2.17 Å / Redundancy: 7.2 % / Mean I/σ(I) obs: 2.8 / Rsym value: 0.268 / % possible all: 98.9 |
| Reflection | *PLUS Num. measured all: 105121 / Rmerge(I) obs: 0.077 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 1NBV Resolution: 2.1→10 Å / Cross valid method: THROUGHOUT / σ(F): 0 Details: OTHER REFINEMENT REMARKS: CONTACTS INVOLVING ZINC ATOMS CAN BE AT DISTANCES LESS THAN THE SPECIFIED DISTANCE CUTOFF OF 2.2 ANGSTROMS FOR CONTACTS NOT NOT INVOLVING HYDROGEN ATOMS. THE ...Details: OTHER REFINEMENT REMARKS: CONTACTS INVOLVING ZINC ATOMS CAN BE AT DISTANCES LESS THAN THE SPECIFIED DISTANCE CUTOFF OF 2.2 ANGSTROMS FOR CONTACTS NOT NOT INVOLVING HYDROGEN ATOMS. THE FOLLOWING ATOMS ARE SITUATED ACROSS A TWO-FOLD AXIS AND HAVE AN OCCUPANCY OF 0.5. IT IS THEREFORE POSSIBLE THAT THESE ATOMS ARE IN CLOSE CONTACT WITH OTHER ATOMS AT DISTANCES LESS THAN THOSE NORMALLY OBSERVED. ATM1 RES C SSEQI ZN ZN L 201 O HOH L 300 O HOH L 303 O HOH L 389 RESIDUE VAL L 56 HAS DIHEDRAL ANGLES WHICH LIE OUTSIDE THEIR EXPECTED RANGE IN THE GAMMA QUADRANT OF THE RAMACHANDRAN PLOT. SOLVENT MOLECULES HOH L 300, HOH L 301 AND HOH L 394 ARE SITUATED ON SPECIAL POSITIONS AND HAVE OCCUPANCIES OF 0.50. RESIDUE VAL L 56 HAS DIHEDRAL ANGLES WHICH LIE OUTSIDE THEIR EXPECTED RANGE IN THE GAMMA QUADRANT OF THE RAMACHANDRAN PLOT.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 19.76 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.1→10 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name: PROLSQ / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS Rfactor all: 0.178 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS |
 Movie
Movie Controller
Controller





















 PDBj
PDBj