[English] 日本語
 Yorodumi
Yorodumi- EMDB-9293: EM structure of Bacillus subtilis ribonucleotide reductase inhibi... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9293 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | EM structure of Bacillus subtilis ribonucleotide reductase inhibited double-helical filament of NrdE alpha subunit with dATP | |||||||||
 Map data Map data | ASU of Bacillus subtilis ribonucleotide reductase inhibited double-helix filament composed of NrdE alpha subunits with dATP | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ribonucleotide reductase / allostery / nucleotide metabolism / filament / dATP / ATP / OXIDOREDUCTASE / PROTEIN FIBRIL | |||||||||
| Function / homology |  Function and homology information Function and homology informationribonucleoside-diphosphate reductase complex / ribonucleoside-diphosphate reductase / ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor / deoxyribonucleotide biosynthetic process / DNA replication / ATP binding Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 6.0 Å | |||||||||
 Authors Authors | Thomas WC / Bacik JP | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2019 Journal: Nat Commun / Year: 2019Title: Convergent allostery in ribonucleotide reductase. Authors: William C Thomas / F Phil Brooks / Audrey A Burnim / John-Paul Bacik / JoAnne Stubbe / Jason T Kaelber / James Z Chen / Nozomi Ando /  Abstract: Ribonucleotide reductases (RNRs) use a conserved radical-based mechanism to catalyze the conversion of ribonucleotides to deoxyribonucleotides. Within the RNR family, class Ib RNRs are notable for ...Ribonucleotide reductases (RNRs) use a conserved radical-based mechanism to catalyze the conversion of ribonucleotides to deoxyribonucleotides. Within the RNR family, class Ib RNRs are notable for being largely restricted to bacteria, including many pathogens, and for lacking an evolutionarily mobile ATP-cone domain that allosterically controls overall activity. In this study, we report the emergence of a distinct and unexpected mechanism of activity regulation in the sole RNR of the model organism Bacillus subtilis. Using a hypothesis-driven structural approach that combines the strengths of small-angle X-ray scattering (SAXS), crystallography, and cryo-electron microscopy (cryo-EM), we describe the reversible interconversion of six unique structures, including a flexible active tetramer and two inhibited helical filaments. These structures reveal the conformational gymnastics necessary for RNR activity and the molecular basis for its control via an evolutionarily convergent form of allostery. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9293.map.gz emd_9293.map.gz | 14.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9293-v30.xml emd-9293-v30.xml emd-9293.xml emd-9293.xml | 20.1 KB 20.1 KB | Display Display |  EMDB header EMDB header |
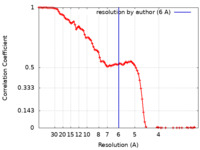
| FSC (resolution estimation) |  emd_9293_fsc.xml emd_9293_fsc.xml | 10.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_9293.png emd_9293.png | 44 KB | ||
| Filedesc metadata |  emd-9293.cif.gz emd-9293.cif.gz | 7 KB | ||
| Others |  emd_9293_half_map_1.map.gz emd_9293_half_map_1.map.gz emd_9293_half_map_2.map.gz emd_9293_half_map_2.map.gz | 59.5 MB 59.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9293 http://ftp.pdbj.org/pub/emdb/structures/EMD-9293 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9293 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9293 | HTTPS FTP |
-Related structure data
| Related structure data |  6myxMC  9272C  6mt9C  6mv9C  6mveC  6mw3C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_9293.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9293.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | ASU of Bacillus subtilis ribonucleotide reductase inhibited double-helix filament composed of NrdE alpha subunits with dATP | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.505 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: Half-set map #1
| File | emd_9293_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-set map #1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
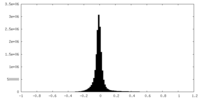
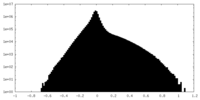
| Density Histograms |
-Half map: Half-set map #2
| File | emd_9293_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-set map #2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
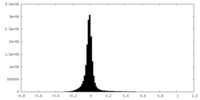
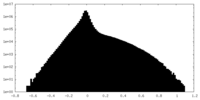
| Density Histograms |
- Sample components
Sample components
-Entire : Inhibited filament of ribonucleoside-diphosphate reductase compos...
| Entire | Name: Inhibited filament of ribonucleoside-diphosphate reductase composed of NrdE alpha subunit |
|---|---|
| Components |
|
-Supramolecule #1: Inhibited filament of ribonucleoside-diphosphate reductase compos...
| Supramolecule | Name: Inhibited filament of ribonucleoside-diphosphate reductase composed of NrdE alpha subunit type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: The filament is a double helix. Each helix is composed of NrdE subunits dimerizing at alternating canonical and non-canonical interfaces. |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Ribonucleoside-diphosphate reductase
| Macromolecule | Name: Ribonucleoside-diphosphate reductase / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO / EC number: ribonucleoside-diphosphate reductase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 80.791469 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSQNQVPKWI QLNNEIMIQK DGKFQFDKDK EAVHSYFVDY INQNTVFFHN LKEKLDYLVE NQYYEEEFLS LYSFEDIKEV FKTAYAKKF RFPSFMSAFK FYNDYALKTN DKKKILERYE DRISIVALFF ANGDTEKAKE YVNLMINQEY QPSTPTFLNA G RKRRGELV ...String: MSQNQVPKWI QLNNEIMIQK DGKFQFDKDK EAVHSYFVDY INQNTVFFHN LKEKLDYLVE NQYYEEEFLS LYSFEDIKEV FKTAYAKKF RFPSFMSAFK FYNDYALKTN DKKKILERYE DRISIVALFF ANGDTEKAKE YVNLMINQEY QPSTPTFLNA G RKRRGELV SCFLLEVNDS LNDISRAIDI SMQLSKLGGG VSLNLSKLRA KGEAIKDVEN ATKGVVGVMK LLDNAFRYAD QM GQRQGSG AAYLNIFHRD INDFLDTKKI SADEDVRVKT LSIGVVIPDK FVELAREDKA AYVFYPHTIY KEYGQHMDEM DMN EMYDKF VDNPRVKKEK INPRKLLEKL AMLRSESGYP YIMFQDNVNK VHANNHISKV KFSNLCSEVL QASQVSSYTD YDEE DEIGL DISCNLGSLN ILNVMEHKSI EKTVKLATDS LTHVSETTDI RNAPAVRRAN KAMKSIGLGA MNLHGYLAQN GIAYE SPEA RDFANTFFMM VNFYSIQRSA EIAKEKGETF DQYEGSTYAT GEYFDKYVST DFSPKYEKIA NLFEGMHIPT TEDWKK LKA FVAEHGMYHS YRLCIAPTGS ISYVQSSTAS VMPIMERIEE RTYGNSKTYY PMPGLASNNW FFYKEAYDMD MFKVVDM IA TIQQHIDQGI SFTLFLKDTM TTRDLNRIDL YAHHRGIKTI YYARTKDTGQ DSCLSCVV UniProtKB: Ribonucleoside-diphosphate reductase |
-Macromolecule #2: 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE
| Macromolecule | Name: 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE / type: ligand / ID: 2 / Number of copies: 8 / Formula: DTP |
|---|---|
| Molecular weight | Theoretical: 491.182 Da |
| Chemical component information |  ChemComp-DTP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 0.81 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.6 Component:
Details: Glycerol in original buffer was diluted to < 0.25% w/v. | |||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 300 K / Instrument: FEI VITROBOT MARK IV / Details: 3.5 seconds blotting. | |||||||||||||||
| Details | Samples of the dATP-induced NrdE filament were produced by incubating 40 uM holo-NrdE with 100 uM dATP and 1 mM CDP in assay buffer. The mixture was then diluted to 10 uM NrdE in the same nucleotide-containing buffer. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Specialist optics | Spherical aberration corrector: None / Chromatic aberration corrector: None |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 4000 pixel / Digitization - Dimensions - Height: 4000 pixel / Digitization - Frames/image: 2-90 / Number grids imaged: 2 / Number real images: 500 / Average exposure time: 20.0 sec. / Average electron dose: 20.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated defocus max: 3.0 µm / Calibrated defocus min: 1.2 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.2 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||
|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: OTHER / Target criteria: Corellation coefficient | ||||||
| Output model |  PDB-6myx: |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)