+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20076 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human TorsinA filament | |||||||||
 Map data Map data | Human TorsinA filament | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | AAA+ ATPase / nucleotide binding / nuclear envelope / endoplasmic reticulum / membrane / HYDROLASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationsynaptic vesicle membrane organization / nuclear membrane organization / nuclear envelope organization / regulation of dopamine uptake involved in synaptic transmission / regulation of protein localization to cell surface / intermediate filament cytoskeleton organization / protein deneddylation / positive regulation of synaptic vesicle endocytosis / misfolded protein binding / wound healing, spreading of cells ...synaptic vesicle membrane organization / nuclear membrane organization / nuclear envelope organization / regulation of dopamine uptake involved in synaptic transmission / regulation of protein localization to cell surface / intermediate filament cytoskeleton organization / protein deneddylation / positive regulation of synaptic vesicle endocytosis / misfolded protein binding / wound healing, spreading of cells / synaptic vesicle transport / kinesin binding / protein localization to nucleus / cytoskeletal protein binding / ERAD pathway / secretory granule / cytoplasmic vesicle membrane / ATP-dependent protein folding chaperone / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / neuron projection development / synaptic vesicle / unfolded protein binding / nuclear envelope / Cargo recognition for clathrin-mediated endocytosis / protein folding / growth cone / response to oxidative stress / nuclear membrane / cytoskeleton / cell adhesion / endoplasmic reticulum lumen / endoplasmic reticulum membrane / endoplasmic reticulum / ATP hydrolysis activity / extracellular exosome / ATP binding / identical protein binding / membrane / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 4.4 Å | |||||||||
 Authors Authors | Zheng W / Demircioglu FE | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2019 Journal: Nat Commun / Year: 2019Title: The AAA + ATPase TorsinA polymerizes into hollow helical tubes with 8.5 subunits per turn. Authors: F Esra Demircioglu / Weili Zheng / Alexander J McQuown / Nolan K Maier / Nicki Watson / Iain M Cheeseman / Vladimir Denic / Edward H Egelman / Thomas U Schwartz /  Abstract: TorsinA is an ER-resident AAA + ATPase, whose deletion of glutamate E303 results in the genetic neuromuscular disease primary dystonia. TorsinA is an unusual AAA + ATPase that needs an ...TorsinA is an ER-resident AAA + ATPase, whose deletion of glutamate E303 results in the genetic neuromuscular disease primary dystonia. TorsinA is an unusual AAA + ATPase that needs an external activator. Also, it likely does not thread a peptide substrate through a narrow central channel, in contrast to its closest structural homologs. Here, we examined the oligomerization of TorsinA to get closer to a molecular understanding of its still enigmatic function. We observe TorsinA to form helical filaments, which we analyzed by cryo-electron microscopy using helical reconstruction. The 4.4 Å structure reveals long hollow tubes with a helical periodicity of 8.5 subunits per turn, and an inner channel of ~ 4 nm diameter. We further show that the protein is able to induce tubulation of membranes in vitro, an observation that may reflect an entirely new characteristic of AAA + ATPases. We discuss the implications of these observations for TorsinA function. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20076.map.gz emd_20076.map.gz | 12.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20076-v30.xml emd-20076-v30.xml emd-20076.xml emd-20076.xml | 11.3 KB 11.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_20076.png emd_20076.png | 218 KB | ||
| Filedesc metadata |  emd-20076.cif.gz emd-20076.cif.gz | 5.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20076 http://ftp.pdbj.org/pub/emdb/structures/EMD-20076 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20076 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20076 | HTTPS FTP |
-Related structure data
| Related structure data |  6oifMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_20076.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20076.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Human TorsinA filament | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.169 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
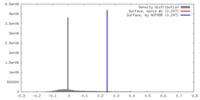
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : TorsinA
| Entire | Name: TorsinA |
|---|---|
| Components |
|
-Supramolecule #1: TorsinA
| Supramolecule | Name: TorsinA / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Torsin-1A
| Macromolecule | Name: Torsin-1A / type: protein_or_peptide / ID: 1 / Number of copies: 25 / Enantiomer: LEVO EC number: Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 32.590248 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GQKRSLSREA LQKDLDDNLF GQHLAKKIIL NAVFGFINNP KPKKPLTLSL HGWTGTGKNF VSKIIAENIY EGGLNSDYVH LFVATLHFP HASNITLYKD QLQLWIRGNV SACARSIFIF DEMDKMHAGL IDAIKPFLDY YDLVDGVSYQ KAMFIFLSNA G AERITDVA ...String: GQKRSLSREA LQKDLDDNLF GQHLAKKIIL NAVFGFINNP KPKKPLTLSL HGWTGTGKNF VSKIIAENIY EGGLNSDYVH LFVATLHFP HASNITLYKD QLQLWIRGNV SACARSIFIF DEMDKMHAGL IDAIKPFLDY YDLVDGVSYQ KAMFIFLSNA G AERITDVA LDFWRSGKQR EDIKLKDIEH ALSVSVFNNK NSGFWHSSLI DRNLIDYFVP FLPLEYKHLK MCIRVEMQSR GY EIDEDIV SRVAEEMTFF PKEERVFSDK GCKTVFTKLD YYYDD UniProtKB: Torsin-1A |
-Macromolecule #2: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 2 / Number of copies: 25 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Grid | Details: unspecified |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Helical parameters - Δz: 5.53 Å Applied symmetry - Helical parameters - Δ&Phi: 42.51 ° Applied symmetry - Helical parameters - Axial symmetry: C1 (asymmetric) Resolution.type: BY AUTHOR / Resolution: 4.4 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: SPIDER / Number images used: 75909 |
|---|---|
| Segment selection | Number selected: 75909 / Software - Name: EMAN2 / Software - details: EMAN2 e2helixboxer.py |
| Startup model | Type of model: NONE / Details: Featureless cylinder |
| Final angle assignment | Type: NOT APPLICABLE / Software - Name: SPIDER |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)