Yorodumi
Yorodumi+ Open data
Open data
 Loading...
Loading...
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6696 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the 90S small subunit pre-ribosome (Mtr4-depleted, Enp1-TAP) | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | ribosome / 90S / pre-ribosome / protein-RNA complex | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of ribosomal protein gene transcription by RNA polymerase II / RNA fragment catabolic process / rRNA small subunit pseudouridine methyltransferase Nep1 / CURI complex / UTP-C complex / rRNA 2'-O-methylation / t-UTP complex / Pwp2p-containing subcomplex of 90S preribosome / nuclear microtubule / Mpp10 complex ...regulation of ribosomal protein gene transcription by RNA polymerase II / RNA fragment catabolic process / rRNA small subunit pseudouridine methyltransferase Nep1 / CURI complex / UTP-C complex / rRNA 2'-O-methylation / t-UTP complex / Pwp2p-containing subcomplex of 90S preribosome / nuclear microtubule / Mpp10 complex / rRNA (pseudouridine) methyltransferase activity / snoRNA guided rRNA 2'-O-methylation / rRNA modification / histone H2AQ104 methyltransferase activity / septum digestion after cytokinesis / regulation of rRNA processing / snRNA binding / RNA folding chaperone / box C/D sno(s)RNA 3'-end processing / rRNA methyltransferase activity / rDNA heterochromatin / endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / regulation of transcription by RNA polymerase I / endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / box C/D methylation guide snoRNP complex / U4/U6 snRNP / tRNA export from nucleus / single-stranded telomeric DNA binding / rRNA primary transcript binding / sno(s)RNA-containing ribonucleoprotein complex / rRNA base methylation / : / U4 snRNA binding / O-methyltransferase activity / SUMOylation of RNA binding proteins / protein localization to nucleolus / rRNA methylation / Protein hydroxylation / U4 snRNP / U3 snoRNA binding / positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay / Formation of the ternary complex, and subsequently, the 43S complex / Translation initiation complex formation / Ribosomal scanning and start codon recognition / snoRNA binding / preribosome, small subunit precursor / precatalytic spliceosome / Major pathway of rRNA processing in the nucleolus and cytosol / establishment of cell polarity / spliceosomal complex assembly / positive regulation of transcription by RNA polymerase I / SRP-dependent cotranslational protein targeting to membrane / GTP hydrolysis and joining of the 60S ribosomal subunit / nucleolar large rRNA transcription by RNA polymerase I / Formation of a pool of free 40S subunits / Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) / Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) / L13a-mediated translational silencing of Ceruloplasmin expression / 90S preribosome / Ub-specific processing proteases / proteasome assembly / regulation of translational fidelity / ribosomal subunit export from nucleus / U4/U6 x U5 tri-snRNP complex / endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / ribosomal small subunit export from nucleus / RNA endonuclease activity / nuclear periphery / Transferases; Transferring one-carbon groups; Methyltransferases / ribosome assembly / maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / spliceosomal complex / maturation of SSU-rRNA / translational initiation / small-subunit processome / enzyme activator activity / mRNA splicing, via spliceosome / maintenance of translational fidelity / cytoplasmic stress granule / rRNA processing / unfolded protein binding / ribosome biogenesis / ribosomal small subunit biogenesis / ribosomal small subunit assembly / small ribosomal subunit rRNA binding / cytosolic small ribosomal subunit / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / cytoplasmic translation / rRNA binding / structural constituent of ribosome / ribosome / translation / mRNA binding / GTPase activity / GTP binding / nucleolus / ATP hydrolysis activity / mitochondrion / RNA binding Similarity search - Function Ribosomal RNA assembly KRR1 / : / : / : / KRR1 small subunit processome component, second KH domain / WD repeat-containing protein 75 second beta-propeller / NOL6/Upt22 / Nrap protein domain 1 / Nrap protein, domain 2 / Nrap protein, domain 3 ...Ribosomal RNA assembly KRR1 / : / : / : / KRR1 small subunit processome component, second KH domain / WD repeat-containing protein 75 second beta-propeller / NOL6/Upt22 / Nrap protein domain 1 / Nrap protein, domain 2 / Nrap protein, domain 3 / Nrap protein, domain 4 / Nrap protein, domain 5 / Nrap protein, domain 6 / Nrap protein domain 1 / Nrap protein PAP/OAS-like domain / Nrap protein domain 3 / Nrap protein nucleotidyltransferase domain 4 / Nrap protein PAP/OAS1-like domain 5 / Nrap protein domain 6 / Ribosomal RNA-processing protein 7, C-terminal domain / Ribosomal RNA-processing protein 7 / Rrp7, RRM-like N-terminal domain / Ribosomal RNA-processing protein 7 (RRP7) C-terminal domain / Rrp7 RRM-like N-terminal domain / rRNA-processing protein Fcf1, PIN domain / WDR3 second beta-propeller domain / WDR3 first beta-propeller domain / U3 small nucleolar RNA-associated protein 18 / U3 small nucleolar RNA-associated protein 13, C-terminal / Periodic tryptophan protein 2 / Utp13 specific WD40 associated domain / BP28, C-terminal domain / Nucleolar protein 58/56, N-terminal / U3 small nucleolar RNA-associated protein 10, N-terminal / U3 small nucleolar RNA-associated protein 10 / : / BP28CT (NUC211) domain / NOP5NT (NUC127) domain / U3 small nucleolar RNA-associated protein 10 / HEATR1-like, HEAT repeats / BP28CT (NUC211) domain / rRNA-processing protein Fcf1/Utp23 / : / Fcf1 / Small-subunit processome, Utp12 / Dip2/Utp12 Family / Small-subunit processome, Utp21 / Utp21 specific WD40 associated putative domain / WDR36/Utp21 second beta-propeller domain / WDR36/Utp21 first beta-propeller / RNA 3'-terminal phosphate cyclase-like, conserved site / Ribosome biogenesis protein Bms1, N-terminal / RNA 3'-terminal phosphate cyclase signature. / Ribosomal RNA-processing protein Rrp9-like / U3 small nucleolar ribonucleoprotein complex, subunit Mpp10 / RNA 3'-terminal phosphate cyclase type 2 / Mpp10 protein / RNA 3'-terminal phosphate cyclase / Ribosomal biogenesis, methyltransferase, EMG1/NEP1 / RNA 3'-terminal phosphate cyclase, insert domain / RNA 3'-terminal phosphate cyclase domain / RNA 3'-terminal phosphate cyclase, insert domain superfamily / RNA 3'-terminal phosphate cyclase domain superfamily / Nucleolar protein Nop56/Nop58 / RNA 3'-terminal phosphate cyclase / EMG1/NEP1 methyltransferase / RNA 3'-terminal phosphate cyclase (RTC), insert domain / rRNA 2'-O-methyltransferase fibrillarin-like / Fibrillarin, conserved site / Fibrillarin / Fibrillarin signature. / Fibrillarin / Krr1, KH1 domain / Krr1 KH1 domain / : / Anaphase-promoting complex subunit 4, WD40 domain / : / Eukaryotic type KH-domain (KH-domain type I) / U3 snoRNP protein/Ribosome production factor 1 / Large family of predicted nucleotide-binding domains / Ribosome biogenesis protein BMS1/TSR1, C-terminal / AARP2CN / Bms1/Tsr1-type G domain / Ribosome biogenesis protein Bms1/Tsr1 / 40S ribosome biogenesis protein Tsr1 and BMS1 C-terminal / AARP2CN (NUC121) domain / Bms1-type guanine nucleotide-binding (G) domain profile. / AARP2CN (NUC121) domain / Protein of unknown function (DUF663) / Anaphase-promoting complex subunit 4 WD40 domain / NOSIC / NOSIC (NUC001) domain / Nop domain / Nop domain superfamily / Nop, C-terminal domain / snoRNA binding domain, fibrillarin / Nop domain profile. / PIN domain / H/ACA ribonucleoprotein complex, subunit Nhp2-like / Brix domain Similarity search - Domain/homology Small ribosomal subunit protein uS4A / Small ribosomal subunit protein uS15 / Small ribosomal subunit protein uS11A / Small ribosomal subunit protein uS8A / Small ribosomal subunit protein uS12A / Small ribosomal subunit protein eS24A / Small ribosomal subunit protein eS30A / Small ribosomal subunit protein eS4A / Small ribosomal subunit protein eS8A / Small ribosomal subunit protein uS17A ...Small ribosomal subunit protein uS4A / Small ribosomal subunit protein uS15 / Small ribosomal subunit protein uS11A / Small ribosomal subunit protein uS8A / Small ribosomal subunit protein uS12A / Small ribosomal subunit protein eS24A / Small ribosomal subunit protein eS30A / Small ribosomal subunit protein eS4A / Small ribosomal subunit protein eS8A / Small ribosomal subunit protein uS17A / Small ribosomal subunit protein uS9A / rRNA 2'-O-methyltransferase fibrillarin / Ribosomal RNA-processing protein 7 / KRR1 small subunit processome component / Periodic tryptophan protein 2 / Small ribosomal subunit protein uS7 / Small ribosomal subunit protein eS7A / U3 small nucleolar ribonucleoprotein protein IMP3 / Small ribosomal subunit protein eS1A / Small ribosomal subunit protein eS27A / Ribosome biogenesis protein UTP30 / 13 kDa ribonucleoprotein-associated protein / U3 small nucleolar RNA-associated protein 18 / U3 small nucleolar RNA-associated protein 10 / U3 small nucleolar RNA-associated protein MPP10 / U3 small nucleolar RNA-associated protein 22 / U3 small nucleolar ribonucleoprotein protein IMP4 / rRNA-processing protein FCF1 / U3 small nucleolar RNA-associated protein 13 / U3 small nucleolar RNA-associated protein 21 / Ribosomal RNA small subunit methyltransferase NEP1 / Ribosomal RNA-processing protein 9 / rRNA processing protein RCL1 / Ribosome biogenesis protein BMS1 / U3 small nucleolar RNA-associated protein 12 / Nucleolar protein 56 / Nucleolar protein 58 / Small ribosomal subunit protein eS28A / Pre-rRNA-processing protein PNO1 Similarity search - Component | |||||||||||||||
| Biological species |   | |||||||||||||||
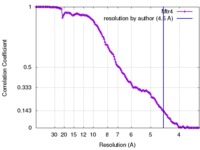
| Method | single particle reconstruction / cryo EM / Resolution: 4.5 Å | |||||||||||||||
 Authors Authors | Ye K / Zhu X | |||||||||||||||
 Citation Citation |  Journal: Elife / Year: 2017 Journal: Elife / Year: 2017Title: Molecular architecture of the 90S small subunit pre-ribosome. Authors: Qi Sun / Xing Zhu / Jia Qi / Weidong An / Pengfei Lan / Dan Tan / Rongchang Chen / Bing Wang / Sanduo Zheng / Cheng Zhang / Xining Chen / Wei Zhang / Jing Chen / Meng-Qiu Dong / Keqiong Ye /  Abstract: Eukaryotic small ribosomal subunits are first assembled into 90S pre-ribosomes. The complete 90S is a gigantic complex with a molecular mass of approximately five megadaltons. Here, we report the ...Eukaryotic small ribosomal subunits are first assembled into 90S pre-ribosomes. The complete 90S is a gigantic complex with a molecular mass of approximately five megadaltons. Here, we report the nearly complete architecture of 90S determined from three cryo-electron microscopy single particle reconstructions at 4.5 to 8.7 angstrom resolution. The majority of the density maps were modeled and assigned to specific RNA and protein components. The nascent ribosome is assembled into isolated native-like substructures that are stabilized by abundant assembly factors. The 5' external transcribed spacer and U3 snoRNA nucleate a large subcomplex that scaffolds the nascent ribosome. U3 binds four sites of pre-rRNA, including a novel site on helix 27 but not the 3' side of the central pseudoknot, and crucially organizes the 90S structure. The 90S model provides significant insight into the principle of small subunit assembly and the function of assembly factors. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6696.map.gz emd_6696.map.gz | 228.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6696-v30.xml emd-6696-v30.xml emd-6696.xml emd-6696.xml | 135 KB 135 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_6696_fsc.xml emd_6696_fsc.xml | 14 KB | Display |  FSC data file FSC data file |
| Images |  emd_6696.png emd_6696.png | 87.7 KB | ||
| Filedesc metadata |  emd-6696.cif.gz emd-6696.cif.gz | 20.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6696 http://ftp.pdbj.org/pub/emdb/structures/EMD-6696 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6696 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6696 | HTTPS FTP |
-Related structure data
| Related structure data |  5wykMC  5ydtM  6695C  6697C  5wwnC  5wwoC  5wxlC  5wxmC  5wy3C  5wyjC  5wylC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_6696.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6696.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.76 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
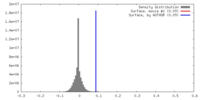
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : 90S small subunit pre-ribosome (Mtr4-depleted, Enp1-TAP)
| Entire | Name: 90S small subunit pre-ribosome (Mtr4-depleted, Enp1-TAP) |
|---|---|
| Components |
|
+Supramolecule #1: 90S small subunit pre-ribosome (Mtr4-depleted, Enp1-TAP)
| Supramolecule | Name: 90S small subunit pre-ribosome (Mtr4-depleted, Enp1-TAP) type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#55 |
|---|---|
| Source (natural) | Organism:  |
+Macromolecule #1: U3 RNA
| Macromolecule | Name: U3 RNA / type: rna / ID: 1 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 106.503258 KDa |
| Sequence | String: GUCGACGUAC UUCAUAGGAU CAUUUCUAUA GGAAUCGUCA CUCUUUGACU CUUCAAAAGA GCCACUGAAU CCAACUUGGU UGAUGAGUC CCAUAACCUU UGUACCCCAG AGUGAGAAAC CGAAAUUGAA UCUAAAUUAG CUUGGUCCGC AAUCCUUAGC G GUUCGGCC ...String: GUCGACGUAC UUCAUAGGAU CAUUUCUAUA GGAAUCGUCA CUCUUUGACU CUUCAAAAGA GCCACUGAAU CCAACUUGGU UGAUGAGUC CCAUAACCUU UGUACCCCAG AGUGAGAAAC CGAAAUUGAA UCUAAAUUAG CUUGGUCCGC AAUCCUUAGC G GUUCGGCC AUCUAUAAUU UUGAAUAAAA AUUUUGCUUU GCCGUUGCAU UUGUAGUUUU UUCCUUUGGA AGUAAUUACA AU AUUUUAU GGCGCGAUGA UCUUGACCCA UCCUAUGUAC UUCUUUUUUG AAGGGAUAGG GCUCUAUGGG UGGGUACAAA UGG CAGUCU GACAAGU |
+Macromolecule #7: 5ETS RNA
| Macromolecule | Name: 5ETS RNA / type: rna / ID: 7 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 225.543094 KDa |
| Sequence | String: AUGCGAAAGC AGUUGAAGAC AAGUUCGAAA AGAGUUUGGA AACGAAUUCG AGUAGGCUUG UCGUUCGUUA UGUUUUUGUA AAUGGCCUC GUCAAACGGU GGAGAGAGUC GCUAGGUGAU CGUCAGAUCU GCCUAGUCUC UAUACAGCGU GUUUAAUUGA C AUGGGUUG ...String: AUGCGAAAGC AGUUGAAGAC AAGUUCGAAA AGAGUUUGGA AACGAAUUCG AGUAGGCUUG UCGUUCGUUA UGUUUUUGUA AAUGGCCUC GUCAAACGGU GGAGAGAGUC GCUAGGUGAU CGUCAGAUCU GCCUAGUCUC UAUACAGCGU GUUUAAUUGA C AUGGGUUG AUGCGUAUUG AGAGAUACAA UUUGGGAAGA AAUUCCCAGA GUGUGUUUCU UUUGCGUUUA ACCUGAACAG UC UCAUCGU GGGCAUCUUG CGAUUCCAUU GGUGAGCAGC GAAGGAUUUG GUGGAUUACU AGCUAAUAGC AAUCUAUUUC AAA GAAUUC AAACUUGGGG GAAUGCCUUG UUGAAUAGCC GGUCGCAAGA CUGUGAUUCU UCAAGUGUAA CCUCCUCUCA AAUC AGCGA UAUCAAACGU ACCAUUCCGU GAAACACCGG GGUAUCUGUU UGGUGGAACC UGAUUAGAGG AAACUCAAAG AGUGC UAUG GUAUGGUGAC GGAGUGCGCU GGUCAAGAGU GUAAAAGCUU UUUGAACAGA GAGCAUUUCC GGCAGCAGAG AGACCU GAA AAAGCAAUUU UUCUGGAAUU UCAGCUGUUU CCAAACUCAA UAAGUAUCUU CUAGCAAGAG GGAAUAGGUG GGAAAAA AA AAAAGAGAUU UCGGUUUCUU UCUUUUUUAC UGCUUGUUGC UUCUUCUUUU AAGAUAGU GENBANK: GENBANK: U53879.1 |
+Macromolecule #32: 18S ribosomal RNA
| Macromolecule | Name: 18S ribosomal RNA / type: rna / ID: 32 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 583.636562 KDa |
| Sequence | String: UAUCUGGUUG AUCCUGCCAG UAGUCAUAUG CUUGUCUCAA AGAUUAAGCC AUGCAUGUCU AAGUAUAAGC AAUUUAUACA GUGAAACUG CGAAUGGCUC AUUAAAUCAG UUAUCGUUUA UUUGAUAGUU CCUUUACUAC AUGGUAUAAC UGUGGUAAUU C UAGAGCUA ...String: UAUCUGGUUG AUCCUGCCAG UAGUCAUAUG CUUGUCUCAA AGAUUAAGCC AUGCAUGUCU AAGUAUAAGC AAUUUAUACA GUGAAACUG CGAAUGGCUC AUUAAAUCAG UUAUCGUUUA UUUGAUAGUU CCUUUACUAC AUGGUAUAAC UGUGGUAAUU C UAGAGCUA AUACAUGCUU AAAAUCUCGA CCCUUUGGAA GAGAUGUAUU UAUUAGAUAA AAAAUCAAUG UCUUCGGACU CU UUGAUGA UUCAUAAUAA CUUUUCGAAU CGCAUGGCCU UGUGCUGGCG AUGGUUCAUU CAAAUUUCUG CCCUAUCAAC UUU CGAUGG UAGGAUAGUG GCCUACCAUG GUUUCAACGG GUAACGGGGA AUAAGGGUUC GAUUCCGGAG AGGGAGCCUG AGAA ACGGC UACCACAUCC AAGGAAGGCA GCAGGCGCGC AAAUUACCCA AUCCUAAUUC AGGGAGGUAG UGACAAUAAA UAACG AUAC AGGGCCCAUU CGGGUCUUGU AAUUGGAAUG AGUACAAUGU AAAUACCUUA ACGAGGAACA AUUGGAGGGC AAGUCU GGU GCCAGCAGCC GCGGUAAUUC CAGCUCCAAU AGCGUAUAUU AAAGUUGUUG CAGUUAAAAA GCUCGUAGUU GAACUUU GG GCCCGGUUGG CCGGUCCGAU UUUUUCGUGU ACUGGAUUUC CAACGGGGCC UUUCCUUCUG GCUAACCUUG AGUCCUUG U GGCUCUUGGC GAACCAGGAC UUUUACUUUG AAAAAAUUAG AGUGUUCAAA GCAGGCGUAU UGCUCGAAUA UAUUAGCAU GGAAUAAUAG AAUAGGACGU UUGGUUCUAU UUUGUUGGUU UCUAGGACCA UCGUAAUGAU UAAUAGGGAC GGUCGGGGGC AUCAGUAUU CAAUUGUCAG AGGUGAAAUU CUUGGAUUUA UUGAAGACUA ACUACUGCGA AAGCAUUUGC CAAGGACGUU U UCAUUAAU CAAGAACGAA AGUUAGGGGA UCGAAGAUGA UCAGAUACCG UCGUAGUCUU AACCAUAAAC UAUGCCGACU AG GGAUCGG GUGGUGUUUU UUUAAUGACC CACUCGGCAC CUUACGAGAA AUCAAAGUCU UUGGGUUCUG GGGGGAGUAU GGU CGCAAG GCUGAAACUU AAAGGAAUUG ACGGAAGGGC ACCACCAGGA GUGGAGCCUG CGGCUUAAUU UGACUCAACA CGGG GAAAC UCACCAGGUC CAGACACAAU AAGGAUUGAC AGAUUGAGAG CUCUUUCUUG AUUUUGUGGG UGGUGGUGCA UGGCC GUUC UUAGUUGGUG GAGUGAUUUG UCUGCUUAAU UGCGAUAACG AACGAGACCU UAACCUACUA AAUAGUGGUG CUAGCA UUU GCUGGUUAUC CACUUCUUAG AGGGACUAUC GGUUUCAAGC CGAUGGAAGU UUGAGGCAAU AACAGGUCUG UGAUGCC CU UAGACGUUCU GGGCCGCACG CGCGCUACAC UGACGGAGCC AGCGAGUCUA ACCUUGGCCG AGAGGUCUUG GUAAUCUU G UGAAACUCCG UCGUGCUGGG GAUAGAGCAU UGUAAUUAUU GCUCUUCAAC GAGGAAUUCC UAGUAAGCGC AAGUCAUCA GCUUGCGUUG AUUACGUCCC UGCCCUUUGU ACACACCGCC CGUCGCUAGU ACCGAUUGAA UGGCUUAGUG AGGCCUCAGG AUCUGCUUA GAGAAGGGGG CAACUCCAUC UCAGAGCGGA GAAUUUGGAC AAACUUGGUC AUUUAGAGGA ACUAAAAGUC G UAACAAGG UUUCCGUAGG UGAACCUGCG GAAGGAUCAU UAAAGAAAUU UAAU GENBANK: GENBANK: CP011558.1 |
+Macromolecule #2: rRNA 2'-O-methyltransferase fibrillarin
| Macromolecule | Name: rRNA 2'-O-methyltransferase fibrillarin / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO EC number: Transferases; Transferring one-carbon groups; Methyltransferases |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 34.525418 KDa |
| Sequence | String: MSFRPGSRGG SRGGSRGGFG GRGGSRGGAR GGSRGGFGGR GGSRGGARGG SRGGFGGRGG SRGGARGGSR GGRGGAAGGA RGGAKVVIE PHRHAGVYIA RGKEDLLVTK NMAPGESVYG EKRISVEEPS KEDGVPPTKV EYRVWNPFRS KLAAGIMGGL D ELFIAPGK ...String: MSFRPGSRGG SRGGSRGGFG GRGGSRGGAR GGSRGGFGGR GGSRGGARGG SRGGFGGRGG SRGGARGGSR GGRGGAAGGA RGGAKVVIE PHRHAGVYIA RGKEDLLVTK NMAPGESVYG EKRISVEEPS KEDGVPPTKV EYRVWNPFRS KLAAGIMGGL D ELFIAPGK KVLYLGAASG TSVSHVSDVV GPEGVVYAVE FSHRPGRELI SMAKKRPNII PIIEDARHPQ KYRMLIGMVD CV FADVAQP DQARIIALNS HMFLKDQGGV VISIKANCID STVDAETVFA REVQKLREER IKPLEQLTLE PYERDHCIVV GRY MRSGLK K UniProtKB: rRNA 2'-O-methyltransferase fibrillarin |
+Macromolecule #3: Nucleolar protein 56
| Macromolecule | Name: Nucleolar protein 56 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 56.961152 KDa |
| Sequence | String: MAPIEYLLFE EPTGYAVFKV KLQQDDIGSR LKEVQEQIND FGAFTKLIEL VSFAPFKGAA EALENANDIS EGLVSESLKA ILDLNLPKA SSKKKNITLA ISDKNLGPSI KEEFPYVDCI SNELAQDLIR GVRLHGEKLF KGLQSGDLER AQLGLGHAYS R AKVKFSVQ ...String: MAPIEYLLFE EPTGYAVFKV KLQQDDIGSR LKEVQEQIND FGAFTKLIEL VSFAPFKGAA EALENANDIS EGLVSESLKA ILDLNLPKA SSKKKNITLA ISDKNLGPSI KEEFPYVDCI SNELAQDLIR GVRLHGEKLF KGLQSGDLER AQLGLGHAYS R AKVKFSVQ KNDNHIIQAI ALLDQLDKDI NTFAMRVKEW YGWHFPELAK LVPDNYTFAK LVLFIKDKAS LNDDSLHDLA AL LNEDSGI AQRVIDNARI SMGQDISETD MENVCVFAQR VASLADYRRQ LYDYLCEKMH TVAPNLSELI GEVIGARLIS HAG SLTNLS KQAASTVQIL GAEKALFRAL KTKGNTPKYG LIYHSGFISK ASAKNKGRIS RYLANKCSMA SRIDNYSEEP SNVF GSVLK KQVEQRLEFY NTGKPTLKNE LAIQEAMELY NKDKPAAEVE ETKEKESSKK RKLEDDDEEK KEKKEKKSKK EKKEK KEKK DKKEKKDKKE KKDKKKKSKD UniProtKB: Nucleolar protein 56 |
+Macromolecule #4: Nucleolar protein 58
| Macromolecule | Name: Nucleolar protein 58 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 57.060344 KDa |
| Sequence | String: MAYVLTETSA GYALLKASDK KIYKSSSLIQ DLDSSDKVLK EFKIAAFSKF NSAANALEEA NSIIEGKVSS QLEKLLEEIK KDKKSTLIV SETKLANAIN KLGLNFNVVS DAVTLDIYRA IKEYLPELLP GMSDNDLSKM SLGLAHSIGR HKLKFSADKV D VMIIQAIA ...String: MAYVLTETSA GYALLKASDK KIYKSSSLIQ DLDSSDKVLK EFKIAAFSKF NSAANALEEA NSIIEGKVSS QLEKLLEEIK KDKKSTLIV SETKLANAIN KLGLNFNVVS DAVTLDIYRA IKEYLPELLP GMSDNDLSKM SLGLAHSIGR HKLKFSADKV D VMIIQAIA LLDDLDKELN TYAMRCKEWY GWHFPELAKI VTDSVAYARI ILTMGIRSKA SETDLSEILP EEIEERVKTA AE VSMGTEI TQTDLDNINA LAEQIVEFAA YREQLSNYLS ARMKAIAPNL TQLVGELVGA RLIAHSGSLI SLAKSPASTI QIL GAEKAL FRALKTKHDT PKYGLLYHAS LVGQATGKNK GKIARVLAAK AAVSLRYDAL AEDRDDSGDI GLESRAKVEN RLSQ LEGRD LRTTPKVVRE AKKVEMTEAR AYNADADTAK AASDSESDSD DEEEEKKEKK EKKRKRDDDE DSKDSKKAKK EKKDK KEKK EKKEKKEKKE KKEKKEKKSK KEKKEKK UniProtKB: Nucleolar protein 58 |
+Macromolecule #5: Ribosomal RNA-processing protein 9
| Macromolecule | Name: Ribosomal RNA-processing protein 9 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 65.146969 KDa |
| Sequence | String: MSDVTQQKKR KRSKGEVNPS KPTVDEEITD PSSNEDEQLE VSDEEDALES EEEFEGENPA DKRRRLAKQY LENLKSEAND ILTDNRNAE EKDLNNLKER TIDEYNNFDA GDLDKDIIAS RLKEDVAEQQ GRVFRYFGDK LLISEAKQSF TRVGENNLTC I SCFQPVLN ...String: MSDVTQQKKR KRSKGEVNPS KPTVDEEITD PSSNEDEQLE VSDEEDALES EEEFEGENPA DKRRRLAKQY LENLKSEAND ILTDNRNAE EKDLNNLKER TIDEYNNFDA GDLDKDIIAS RLKEDVAEQQ GRVFRYFGDK LLISEAKQSF TRVGENNLTC I SCFQPVLN KYTFEESSNG DKNKGRLFAY TVSKDLQLTK YDITDFSKRP KKLKYAKGGA KYIPTSKHEY ENTTEGHYDE IL TVAASPD GKYVVTGGRD RKLIVWSTES LSPVKVIPTK DRRGEVLSLA FRKNSDQLYA SCADFKIRTY SINQFSQLEI LYG HHDIVE DISALAMERC VTVGARDRTA MLWKIPDETR LTFRGGDEPQ KLLRRWMKEN AKEGEDGEVK YPDESEAPLF FCEG SIDVV SMVDDFHFIT GSDNGNICLW SLAKKKPIFT ERIAHGILPE PSFNDISGET DEELRKRQLQ GKKLLQPFWI TSLYA IPYS NVFISGSWSG SLKVWKISDN LRSFELLGEL SGAKGVVTKI QVVESGKHGK EKFRILASIA KEHRLGRWIA NVSGAR NGI YSAVIDQTGF UniProtKB: Ribosomal RNA-processing protein 9 |
+Macromolecule #6: 13 kDa ribonucleoprotein-associated protein
| Macromolecule | Name: 13 kDa ribonucleoprotein-associated protein / type: protein_or_peptide / ID: 6 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 13.582855 KDa |
| Sequence | String: MSAPNPKAFP LADAALTQQI LDVVQQAANL RQLKKGANEA TKTLNRGISE FIIMAADCEP IEILLHLPLL CEDKNVPYVF VPSRVALGR ACGVSRPVIA ASITTNDASA IKTQIYAVKD KIETLLI UniProtKB: 13 kDa ribonucleoprotein-associated protein |
+Macromolecule #8: Utp4
| Macromolecule | Name: Utp4 / type: protein_or_peptide / ID: 8 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 66.059172 KDa |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) ...String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) |
+Macromolecule #9: Utp5
| Macromolecule | Name: Utp5 / type: protein_or_peptide / ID: 9 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 54.740609 KDa |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) ...String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) |
+Macromolecule #10: Utp8
| Macromolecule | Name: Utp8 / type: protein_or_peptide / ID: 10 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 60.69775 KDa |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) ...String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) |
+Macromolecule #11: Utp9
| Macromolecule | Name: Utp9 / type: protein_or_peptide / ID: 11 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 48.953438 KDa |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) ...String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) |
+Macromolecule #12: U3 small nucleolar RNA-associated protein 10
| Macromolecule | Name: U3 small nucleolar RNA-associated protein 10 / type: protein_or_peptide / ID: 12 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 200.298984 KDa |
| Sequence | String: MSSLSDQLAQ VASNNATVAL DRKRRQKLHS ASLIYNSKTA ATQDYDFIFE NASKALEELS QIEPKFAIFS RTLFSESSIS LDRNVQTKE EIKDLDNAIN AYLLLASSKW YLAPTLHATE WLVRRFQIHV KNTEMLLLST LNYYQTPVFK RILSIIKLPP L FNCLSNFV ...String: MSSLSDQLAQ VASNNATVAL DRKRRQKLHS ASLIYNSKTA ATQDYDFIFE NASKALEELS QIEPKFAIFS RTLFSESSIS LDRNVQTKE EIKDLDNAIN AYLLLASSKW YLAPTLHATE WLVRRFQIHV KNTEMLLLST LNYYQTPVFK RILSIIKLPP L FNCLSNFV RSEKPPTALT MIKLFNDMDF LKLYTSYLDQ CIKHNATYTN QLLFTTCCFI NVVAFNSNND EKLNQLVPIL LE ISAKLLA SKSKDCQIAA HTILVVFATA LPLKKTIILA AMETILSNLD AKEAKHSALL TICKLFQTLK GQGNVDQLPS KIF KLFDSK FDTVSILTFL DKEDKPVCDK FITSYTRSIA RYDRSKLNII LSLLKKIRLE RYEVRLIITD LIYLSEILED KSQL VELFE YFISINEDLV LKCLKSLGLT GELFEIRLTT SLFTNADVNT DIVKQLSDPV ETTKKDTASF QTFLDKHSEL INTTN VSML TETGERYKKV LSLFTEAIGK GYKASSFLTS FFTTLESRIT FLLRVTISPA APTALKLISL NNIAKYINSI EKEVNI FTL VPCLICALRD ASIKVRTGVK KILSLIAKRP STKHYFLSDK LYGENVTIPM LNPKDSEAWL SGFLNEYVTE NYDISRI LT PKRNEKVFLM FWANQALLIP SPYAKTVLLD NLNKSPTYAS SYSSLFEEFI SHYLENRSSW EKSCIANKTN FEHFERSL V NLVSPKEKQS FMIDFVLSAL NSDYEQLANI AAERLISIFA SLNNAQKLKI VQNIVDSSSN VESSYDTVGV LQSLPLDSD IFVSILNQNS ISNEMDQTDF SKRRRRRSST SKNAFLKEEV SQLAELHLRK LTIILEALDK VRNVGSEKLL FTLLSLLSDL ETLDQDGGL PVLYAQETLI SCTLNTITYL KEHGCTELTN VRADILVSAI RNSASPQVQN KLLLVIGSLA TLSSEVILHS V MPIFTFMG AHSIRQDDEF TTKVVERTIL TVVPALIKNS KGNEKEEMEF LLLSFTTALQ HVPRHRRVKL FSTLIKTLDP VK ALGSFLF LIAQQYSSAL VNFKIGEARI LIEFIKALLV DLHVNEELSG LNDLLDIIKL LTSSKSSSEK KKSLESRVLF SNG VLNFSE SEFLTFMNNT FEFINKITEE TDQDYYDVRR NLRLKVYSVL LDETSDKKLI RNIREEFGTL LEGVLFFINS VELT FSCIT SQENEEASDS ETSLSDHTTE IKEILFKVLG NVLQILPVDE FVNAVLPLLS TSTNEDIRYH LTLVIGSKFE LEGSE AIPI VNNVMKVLLD RMPLESKSVV ISQVILNTMT ALVSKYGKKL EGSILTQALT LATEKVSSDM TEVKISSLAL ITNCVQ VLG VKSIAFYPKI VPPSIKLFDA SLADSSNPLK EQLQVAILLL FAGLIKRIPS FLMSNILDVL HVIYFSREVD SSIRLSV IS LIIENIDLKE VLKVLFRIWS TEIATSNDTV AVSLFLSTLE STVENIDKKS ATSQSPIFFK LLLSLFEFRS ISSFDNNT I SRIEASVHEI SNSYVLKMND KVFRPLFVIL VRWAFDGEGV TNAGITETER LLAFFKFFNK LQENLRGIIT SYFTYLLEP VDMLLKRFIS KDMENVNLRR LVINSLTSSL KFDRDEYWKS TSRFELISVS LVNQLSNIEN SIGKYLVKAI GALASNNSGV DEHNQILNK LIVEHMKASC SSNEKLWAIR AMKLIYSKIG ESWLVLLPQL VPVIAELLED DDEEIEREVR TGLVKVVENV L GEPFDRYL D UniProtKB: U3 small nucleolar RNA-associated protein 10 |
+Macromolecule #13: Utp15
| Macromolecule | Name: Utp15 / type: protein_or_peptide / ID: 13 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 43.676871 KDa |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) ...String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK) |
+Macromolecule #14: Utp17
| Macromolecule | Name: Utp17 / type: protein_or_peptide / ID: 14 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 76.271414 KDa |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) ...String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) |
+Macromolecule #15: Ribosome biogenesis protein BMS1
| Macromolecule | Name: Ribosome biogenesis protein BMS1 / type: protein_or_peptide / ID: 15 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 135.792281 KDa |
| Sequence | String: MEQSNKQHRK AKEKNTAKKK LHTQGHNAKA FAVAAPGKMA RTMQRSSDVN ERKLHVPMVD RTPEDDPPPF IVAVVGPPGT GKTTLIRSL VRRMTKSTLN DIQGPITVVS GKHRRLTFLE CPADDLNAMI DIAKIADLVL LLIDGNFGFE METMEFLNIA Q HHGMPRVL ...String: MEQSNKQHRK AKEKNTAKKK LHTQGHNAKA FAVAAPGKMA RTMQRSSDVN ERKLHVPMVD RTPEDDPPPF IVAVVGPPGT GKTTLIRSL VRRMTKSTLN DIQGPITVVS GKHRRLTFLE CPADDLNAMI DIAKIADLVL LLIDGNFGFE METMEFLNIA Q HHGMPRVL GVATHLDLFK SQSTLRASKK RLKHRFWTEV YQGAKLFYLS GVINGRYPDR EILNLSRFIS VMKFRPLKWR NE HPYMLAD RFTDLTHPEL IETQGLQIDR KVAIYGYLHG TPLPSAPGTR VHIAGVGDFS VAQIEKLPDP CPTPFYQQKL DDF EREKMK EEAKANGEIT TASTTRRRKR LDDKDKLIYA PMSDVGGVLM DKDAVYIDIG KKNEEPSFVP GQERGEGEKL MTGL QSVEQ SIAEKFDGVG LQLFSNGTEL HEVADHEGMD VESGEESIED DEGKSKGRTS LRKPRIYGKP VQEEDADIDN LPSDE EPYT NDDDVQDSEP RMVEIDFNNT GEQGAEKLAL ETDSEFEESE DEFSWERTAA NKLKKTESKK RTWNIGKLIY MDNISP EEC IRRWRGEDDD SKDESDIEED VDDDFFRKKD GTVTKEGNKD HAVDLEKFVP YFDTFEKLAK KWKSVDAIKE RFLGAGI LG NDNKTKSDSN EGGEELYGDF EDLEDGNPSE QAEDNSDKES EDEDENEDTN GDDDNSFTNF DAEEKKDLTM EQEREMNA A KKEKLRAQFE IEEGENFKED DENNEYDTWY ELQKAKISKQ LEINNIEYQE MTPEQRQRIE GFKAGSYVRI VFEKVPMEF VKNFNPKFPI VMGGLLPTEI KFGIVKARLR RHRWHKKILK TNDPLVLSLG WRRFQTLPIY TTTDSRTRTR MLKYTPEHTY CNAAFYGPL CSPNTPFCGV QIVANSDTGN GFRIAATGIV EEIDVNIEIV KKLKLVGFPY KIFKNTAFIK DMFSSAMEVA R FEGAQIKT VSGIRGEIKR ALSKPEGHYR AAFEDKILMS DIVILRSWYP VRVKKFYNPV TSLLLKEKTE WKGLRLTGQI RA AMNLETP SNPDSAYHKI ERVERHFNGL KVPKAVQKEL PFKSQIHQMK PQKKKTYMAK RAVVLGGDEK KARSFIQKVL TIS KAKDSK RKEQKASQRK ERLKKLAKME EEKSQRDKEK KKEYFAQNGK RTTMGGDDES RPRKMRR UniProtKB: Ribosome biogenesis protein BMS1 |
+Macromolecule #16: Periodic tryptophan protein 2
| Macromolecule | Name: Periodic tryptophan protein 2 / type: protein_or_peptide / ID: 16 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 104.097039 KDa |
| Sequence | String: MKSDFKFSNL LGTVYRQGNI TFSDDGKQLL SPVGNRVSVF DLINNKSFTF EYEHRKNIAA IDLNKQGTLL ISIDEDGRAI LVNFKARNV LHHFNFKEKC SAVKFSPDGR LFALASGRFL QIWKTPDVNK DRQFAPFVRH RVHAGHFQDI TSLTWSQDSR F ILTTSKDL ...String: MKSDFKFSNL LGTVYRQGNI TFSDDGKQLL SPVGNRVSVF DLINNKSFTF EYEHRKNIAA IDLNKQGTLL ISIDEDGRAI LVNFKARNV LHHFNFKEKC SAVKFSPDGR LFALASGRFL QIWKTPDVNK DRQFAPFVRH RVHAGHFQDI TSLTWSQDSR F ILTTSKDL SAKIWSVDSE EKNLAATTFN GHRDYVMGAF FSHDQEKIYT VSKDGAVFVW EFTKRPSDDD DNESEDDDKQ EE VDISKYS WRITKKHFFY ANQAKVKCVT FHPATRLLAV GFTSGEFRLY DLPDFTLIQQ LSMGQNPVNT VSVNQTGEWL AFG SSKLGQ LLVYEWQSES YILKQQGHFD STNSLAYSPD GSRVVTASED GKIKVWDITS GFCLATFEEH TSSVTAVQFA KRGQ VMFSS SLDGTVRAWD LIRYRNFRTF TGTERIQFNC LAVDPSGEVV CAGSLDNFDI HVWSVQTGQL LDALSGHEGP VSCLS FSQE NSVLASASWD KTIRIWSIFG RSQQVEPIEV YSDVLALSMR PDGKEVAVST LKGQISIFNI EDAKQVGNID CRKDII SGR FNQDRFTAKN SERSKFFTTI HYSFDGMAIV AGGNNNSICL YDVPNEVLLK RFIVSRNMAL NGTLEFLNSK KMTEAGS LD LIDDAGENSD LEDRIDNSLP GSQRGGDLST RKMRPEVRVT SVQFSPTANA FAAASTEGLL IYSTNDTILF DPFDLDVD V TPHSTVEALR EKQFLNALVM AFRLNEEYLI NKVYEAIPIK EIPLVASNIP AIYLPRILKF IGDFAIESQH IEFNLIWIK ALLSASGGYI NEHKYLFSTA MRSIQRFIVR VAKEVVNTTT DNKYTYRFLV STDGSMEDGA ADDDEVLLKD DADEDNEENE ENDVVMESD DEEGWIGFNG KDNKLPLSNE NDSSDEEENE KELP UniProtKB: Periodic tryptophan protein 2 |
+Macromolecule #17: U3 small nucleolar RNA-associated protein 12
| Macromolecule | Name: U3 small nucleolar RNA-associated protein 12 / type: protein_or_peptide / ID: 17 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 106.481133 KDa |
| Sequence | String: MVKSYQRFEQ AAAFGVIASN ANCVWIPASS GNSNGSGPGQ LITSALEDVN IWDIKTGDLV SKLSDGLPPG ASDARGAKPA ECTYLEAHK DTDLLAVGYA DGVIKVWDLM SKTVLLNFNG HKAAITLLQF DGTGTRLISG SKDSNIIVWD LVGEVGLYKL R SHKDSITG ...String: MVKSYQRFEQ AAAFGVIASN ANCVWIPASS GNSNGSGPGQ LITSALEDVN IWDIKTGDLV SKLSDGLPPG ASDARGAKPA ECTYLEAHK DTDLLAVGYA DGVIKVWDLM SKTVLLNFNG HKAAITLLQF DGTGTRLISG SKDSNIIVWD LVGEVGLYKL R SHKDSITG FWCQGEDWLI STSKDGMIKL WDLKTHQCIE THIAHTGECW GLAVKDDLLI TTGTDSQVKI WKLDIENDKM GG KLTEMGI FEKQSKQRGL KIEFITNSSD KTSFFYIQNA DKTIETFRIR KEEEIARGLK KREKRLKEKG LTEEEIAKSI KES YSSFIL HPFQTIRSLY KIKSASWTTV SSSKLELVLT TSSNTIEYYS IPYEKRDPTS PAPLKTHTIE LQGQRTDVRS IDIS DDNKL LATASNGSLK IWNIKTHKCI RTFECGYALT CKFLPGGLLV ILGTRNGELQ LFDLASSSLL DTIEDAHDAA IWSLD LTSD GKRLVTGSAD KTVKFWDFKV ENSLVPGTKN KFLPVLKLHH DTTLELTDDI LCVRVSPDDR YLAISLLDNT VKVFFL DSM KFYLSLYGHK LPVLSIDISF DSKMIITSSA DKNIKIWGLD FGDCHKSLFA HQDSIMNVKF LPQSHNFFSC SKDAVVK YW DGEKFECIQK LYAHQSEVWA LAVATDGGFV VSSSHDHSIR IWEETEDQVF LEEEKEKELE EQYEDTLLTS LEEGNGDD A FKADASGEGV EDEASGVHKQ TLESLKAGER LMEALDLGIA EIEGLEAYNR DMKLWQRKKL GEAPIKPQGN AVLIAVNKT PEQYIMDTLL RIRMSQLEDA LMVMPFSYVL KFLKFIDTVM QNKTLLHSHL PLICKNLFFI IKFNHKELVS QKNEELKLQI NRVKTELRS ALKSTEDDLG FNVQGLKFVK QQWNLRHNYE FVDEYDQQEK ESNSARKRVF GTVI UniProtKB: U3 small nucleolar RNA-associated protein 12 |
+Macromolecule #18: U3 small nucleolar RNA-associated protein 13
| Macromolecule | Name: U3 small nucleolar RNA-associated protein 13 / type: protein_or_peptide / ID: 18 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 91.132562 KDa |
| Sequence | String: MDLKTSYKGI SLNPIYAGSS AVATVSENGK ILATPVLDEI NIIDLTPGSR KILHKISNED EQEITALKLT PDGQYLTYVS QAQLLKIFH LKTGKVVRSM KISSPSYILD ADSTSTLLAV GGTDGSIIVV DIENGYITHS FKGHGGTISS LKFYGQLNSK I WLLASGDT ...String: MDLKTSYKGI SLNPIYAGSS AVATVSENGK ILATPVLDEI NIIDLTPGSR KILHKISNED EQEITALKLT PDGQYLTYVS QAQLLKIFH LKTGKVVRSM KISSPSYILD ADSTSTLLAV GGTDGSIIVV DIENGYITHS FKGHGGTISS LKFYGQLNSK I WLLASGDT NGMVKVWDLV KRKCLHTLQE HTSAVRGLDI IEVPDNDEPS LNLLSGGRDD IINLWDFNMK KKCKLLKTLP VN QQVESCG FLKDGDGKRI IYTAGGDAIF QLIDSESGSV LKRTNKPIEE LFIIGVLPIL SNSQMFLVLS DQTLQLINVE EDL KNDEDT IQVTSSIAGN HGIIADMRYV GPELNKLALA TNSPSLRIIP VPDLSGPEAS LPLDVEIYEG HEDLLNSLDA TEDG LWIAT ASKDNTAIVW RYNENSCKFD IYAKYIGHSA AVTAVGLPNI VSKGYPEFLL TASNDLTIKK WIIPKPTASM DVQII KVSE YTRHAHEKDI NALSVSPNDS IFATASYDKT CKIWNLENGE LEATLANHKR GLWDVSFCQY DKLLATSSGD KTVKIW SLD TFSVMKTLEG HTNAVQRCSF INKQKQLISC GADGLIKIWD CSSGECLKTL DGHNNRLWAL STMNDGDMIV SADADGV FQ FWKDCTEQEI EEEQEKAKLQ VEQEQSLQNY MSKGDWTNAF LLAMTLDHPM RLFNVLKRAL GESRSRQDTE EGKIEVIF N EELDQAISIL NDEQLILLMK RCRDWNTNAK THTIAQRTIR CILMHHNIAK LSEIPGMVKI VDAIIPYTQR HFTRVDNLV EQSYILDYAL VEMDKLF UniProtKB: U3 small nucleolar RNA-associated protein 13 |
+Macromolecule #19: U3 small nucleolar RNA-associated protein 18
| Macromolecule | Name: U3 small nucleolar RNA-associated protein 18 / type: protein_or_peptide / ID: 19 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 66.49425 KDa |
| Sequence | String: MTMATTAMNV SVPPPDEEEQ LLAKFVFGDT TDLQENLAKF NADFIFNEQE MDVEDQEDEG SESDNSEEDE AQNGELDHVN NDQLFFVDD GGNEDSQDKN EDTMDVDDED DSSSDDYSED SEEAAWIDSD DEKIKVPILV TNKTKKLRTS YNESKINGVH Y INRLRSQF ...String: MTMATTAMNV SVPPPDEEEQ LLAKFVFGDT TDLQENLAKF NADFIFNEQE MDVEDQEDEG SESDNSEEDE AQNGELDHVN NDQLFFVDD GGNEDSQDKN EDTMDVDDED DSSSDDYSED SEEAAWIDSD DEKIKVPILV TNKTKKLRTS YNESKINGVH Y INRLRSQF EKIYPRPKWV DDESDSELDD EEDDEEEGSN NVINGDINAL TKILSTTYNY KDTLSNSKLL PPKKLDIVRL KD ANASHPS HSAIQSLSFH PSKPLLLTGG YDKTLRIYHI DGKTNHLVTS LHLVGSPIQT CTFYTSLSNQ NQQNIFTAGR RRY MHSWDL SLENLTHSQT AKIEKFSRLY GHESTQRSFE NFKVAHLQNS QTNSVHGIVL LQGNNGWINI LHSTSGLWLM GCKI EGVIT DFCIDYQPIS RGKFRTILIA VNAYGEVWEF DLNKNGHVIR RWKDQGGVGI TKIQVGGGTT TTCPALQISK IKQNR WLAV GSESGFVNLY DRNNAMTSST PTPVAALDQL TTTISNLQFS PDGQILCMAS RAVKDALRLV HLPSCSVFSN WPTSGT PLG KVTSVAFSPS GGLLAVGNEQ GKVRLWKLNH Y UniProtKB: U3 small nucleolar RNA-associated protein 18 |
+Macromolecule #20: U3 small nucleolar RNA-associated protein 21
| Macromolecule | Name: U3 small nucleolar RNA-associated protein 21 / type: protein_or_peptide / ID: 20 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 104.927844 KDa |
| Sequence | String: MSIDLKKRKV EEDVRSRGKN SKIFSPFRII GNVSNGVPFA TGTLGSTFYI VTCVGKTFQI YDANTLHLLF VSEKETPSSI VALSAHFHY VYAAYENKVG IYKRGIEEHL LELETDANVE HLCIFGDYLC ASTDDNSIFI YKKSDPQDKY PSEFYTKLTV T EIQGGEIV ...String: MSIDLKKRKV EEDVRSRGKN SKIFSPFRII GNVSNGVPFA TGTLGSTFYI VTCVGKTFQI YDANTLHLLF VSEKETPSSI VALSAHFHY VYAAYENKVG IYKRGIEEHL LELETDANVE HLCIFGDYLC ASTDDNSIFI YKKSDPQDKY PSEFYTKLTV T EIQGGEIV SLQHLATYLN KLTVVTKSNV LLFNVRTGKL VFTSNEFPDQ ITTAEPAPVL DIIALGTVTG EVIMFNMRKG KR IRTIKIP QSRISSLSFR TDGSSHLSVG TSSGDLIFYD LDRRSRIHVL KNIHRESYGG VTQATFLNGQ PIIVTSGGDN SLK EYVFDP SLSQGSGDVV VQPPRYLRSR GGHSQPPSYI AFADSQSHFM LSASKDRSLW SFSLRKDAQS QEMSQRLHKK QDGG RVGGS TIKSKFPEIV ALAIENARIG EWENIITAHK DEKFARTWDM RNKRVGRWTF DTTDDGFVKS VAMSQCGNFG FIGSS NGSI TIYNMQSGIL RKKYKLHKRA VTGISLDGMN RKMVSCGLDG IVGFYDFNKS TLLGKLKLDA PITAMVYHRS SDLFAL ALD DLSIVVIDAV TQRVVRQLWG HSNRITAFDF SPEGRWIVSA SLDSTIRTWD LPTGGCIDGI IVDNVATNVK FSPNGDL LA TTHVTGNGIC IWTNRAQFKT VSTRTIDESE FARMALPSTS VRGNDSMLSG ALESNGGEDL NDIDFNTYTS LEQIDKEL L TLSIGPRSKM NTLLHLDVIR KRSKPKEAPK KSEKLPFFLQ LSGEKVGDEA SVREGIAHET PEEIHRRDQE AQKKLDAEE QMNKFKVTGR LGFESHFTKQ LREGSQSKDY SSLLATLINF SPAAVDLEIR SLNSFEPFDE IVWFIDALTQ GLKSNKNFEL YETFMSLLF KAHGDVIHAN NKNQDIASAL QNWEDVHKKE DRLDDLVKFC MGVAAFVTTA UniProtKB: U3 small nucleolar RNA-associated protein 21 |
+Macromolecule #21: Ribosomal RNA-processing protein 7
| Macromolecule | Name: Ribosomal RNA-processing protein 7 / type: protein_or_peptide / ID: 21 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 34.526441 KDa |
| Sequence | String: MGIEDISAMK NGFIVVPFKL PDHKALPKSQ EASLHFMFAK RHQSSNSNES DCLFLVNLPL LSNIEHMKKF VGQLCGKYDT VSHVEELLY NDEFGLHEVD LSALTSDLMS STDVNEKRYT PRNTALLKFV DAASINNCWN ALKKYSNLHA KHPNELFEWT Y TTPSFTTF ...String: MGIEDISAMK NGFIVVPFKL PDHKALPKSQ EASLHFMFAK RHQSSNSNES DCLFLVNLPL LSNIEHMKKF VGQLCGKYDT VSHVEELLY NDEFGLHEVD LSALTSDLMS STDVNEKRYT PRNTALLKFV DAASINNCWN ALKKYSNLHA KHPNELFEWT Y TTPSFTTF VNFYKPLDID YLKEDIHTHM AIFEQREAQA QEDVQSSIVD EDGFTLVVGK NTKSLNSIRK KILNKNPLSK HE NKAKPIS NIDKKAKKDF YRFQVRERKK QEINQLLSKF KEDQERIKVM KAKRKFNPYT UniProtKB: Ribosomal RNA-processing protein 7 |
+Macromolecule #22: U3 small nucleolar RNA-associated protein 22
| Macromolecule | Name: U3 small nucleolar RNA-associated protein 22 / type: protein_or_peptide / ID: 22 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 140.660141 KDa |
| Sequence | String: MATSVKRKAS ETSDQNIVKV QKKHSTQDST TDNGSKENDH SSQAINERTV PEQENDESDT SPESNEVATN TAATRHNGKV TATESYDIH IARETAELFK SNIFKLQIDE LLEQVKLKQK HVLKVEKFLH KLYDILQEIP DWEEKSLAEV DSFFKNKIVS V PFVDPKPI ...String: MATSVKRKAS ETSDQNIVKV QKKHSTQDST TDNGSKENDH SSQAINERTV PEQENDESDT SPESNEVATN TAATRHNGKV TATESYDIH IARETAELFK SNIFKLQIDE LLEQVKLKQK HVLKVEKFLH KLYDILQEIP DWEEKSLAEV DSFFKNKIVS V PFVDPKPI PQNTNYKFNY KKPDISLIGS FALKAGIYQP NGSSIDTLLT MPKELFEKKD FLNFRCLHKR SVYLAYLTHH LL ILLKKDK LDSFLQLEYS YFDNDPLLPI LRISCSKPTG DSLSDYNFYK TRFSINLLIG FPYKVFEPKK LLPNRNCIRI AQE SKEQSL PATPLYNFSV LSSSTHENYL KYLYKTKKQT ESFVEATVLG RLWLQQRGFS SNMSHSGSLG GFGTFEFTIL MAAL LNGGG INSNKILLHG FSSYQLFKGV IKYLATMDLC HDGHLQFHSN PENSSSSPAS KYIDEGFQTP TLFDKSTKVN ILTKM TVSS YQILKEYAGE TLRMLNNVVQ DQFSNIFLTN ISRFDNLKYD LCYDVQLPLG KYNNLETSLA ATFGSMERVK FITLEN FLA HKITNVARYA LGDRIKYIQI EMVGQKSDFP ITKRKVYSNT GGNHFNFDFV RVKLIVNPSE CDKLVTKGPA HSETMST EA AVFKNFWGIK SSLRRFKDGS ITHCCVWSTS SSEPIISSIV NFALQKHVSK KAQISNETIK KFHNFLPLPN LPSSAKTS V LNLSSFFNLK KSFDDLYKII FQMKLPLSVK SILPVGSAFR YTSLCQPVPF AYSDPDFFQD VILEFETSPK WPDEITSLE KAKTAFLLKI QEELSANSST YRSFFSRDES IPYNLEIVTL NILTPEGYGF KFRVLTERDE ILYLRAIANA RNELKPELEA TFLKFTAKY LASVRHTRTL ENISHSYQFY SPVVRLFKRW LDTHLLLGHI TDELAELIAI KPFVDPAPYF IPGSLENGFL K VLKFISQW NWKDDPLILD LVKPEDDIRD TFETSIGAGS ELDSKTMKKL SERLTLAQYK GIQMNFTNLR NSDPNGTHLQ FF VASKNDP SGILYSSGIP LPIATRLTAL AKVAVNLLQT HGLNQQTINL LFTPGLKDYD FVVDLRTPIG LKSSCGILSA TEF KNITND QAPSNFPENL NDLSEKMDPT YQLVKYLNLK YKNSLILSSR KYIGVNGGEK GDKNVITGLI KPLFKGAHKF RVNL DCNVK PVDDENVILN KEAIFHEIAA FGNDMVINFE TD UniProtKB: U3 small nucleolar RNA-associated protein 22 |
+Macromolecule #23: Ribosomal RNA small subunit methyltransferase NEP1
| Macromolecule | Name: Ribosomal RNA small subunit methyltransferase NEP1 / type: protein_or_peptide / ID: 23 / Number of copies: 2 / Enantiomer: LEVO EC number: rRNA small subunit pseudouridine methyltransferase Nep1 |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 27.936461 KDa |
| Sequence | String: MVEDSRVRDA LKGGDQKALP ASLVPQAPPV LTSKDKITKR MIVVLAMASL ETHKISSNGP GGDKYVLLNC DDHQGLLKKM GRDISEARP DITHQCLLTL LDSPINKAGK LQVYIQTSRG ILIEVNPTVR IPRTFKRFSG LMVQLLHKLS IRSVNSEEKL L KVIKNPIT ...String: MVEDSRVRDA LKGGDQKALP ASLVPQAPPV LTSKDKITKR MIVVLAMASL ETHKISSNGP GGDKYVLLNC DDHQGLLKKM GRDISEARP DITHQCLLTL LDSPINKAGK LQVYIQTSRG ILIEVNPTVR IPRTFKRFSG LMVQLLHKLS IRSVNSEEKL L KVIKNPIT DHLPTKCRKV TLSFDAPVIR VQDYIEKLDD DESICVFVGA MARGKDNFAD EYVDEKVGLS NYPLSASVAC SK FCHGAED AWNIL UniProtKB: Ribosomal RNA small subunit methyltransferase NEP1 |
+Macromolecule #24: Enp2
| Macromolecule | Name: Enp2 / type: protein_or_peptide / ID: 24 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 60.187137 KDa |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) ...String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK) |
+Macromolecule #25: KRR1 small subunit processome component
| Macromolecule | Name: KRR1 small subunit processome component / type: protein_or_peptide / ID: 25 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 37.226254 KDa |
| Sequence | String: MVSTHNRDKP WDTDDIDKWK IEEFKEEDNA SGQPFAEESS FMTLFPKYRE SYLKTIWNDV TRALDKHNIA CVLDLVEGSM TVKTTRKTY DPAIILKARD LIKLLARSVP FPQAVKILQD DMACDVIKIG NFVTNKERFV KRRQRLVGPN GNTLKALELL T KCYILVQG ...String: MVSTHNRDKP WDTDDIDKWK IEEFKEEDNA SGQPFAEESS FMTLFPKYRE SYLKTIWNDV TRALDKHNIA CVLDLVEGSM TVKTTRKTY DPAIILKARD LIKLLARSVP FPQAVKILQD DMACDVIKIG NFVTNKERFV KRRQRLVGPN GNTLKALELL T KCYILVQG NTVSAMGPFK GLKEVRRVVE DCMKNIHPIY HIKELMIKRE LAKRPELANE DWSRFLPMFK KRNVARKKPK KI RNVEKKV YTPFPPAQLP RKVDLEIESG EYFLSKREKQ MKKLNEQKEK QMEREIERQE ERAKDFIAPE EEAYKPNQN UniProtKB: KRR1 small subunit processome component |
+Macromolecule #26: U3 small nucleolar ribonucleoprotein protein IMP3
| Macromolecule | Name: U3 small nucleolar ribonucleoprotein protein IMP3 / type: protein_or_peptide / ID: 26 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 21.928529 KDa |
| Sequence | String: MVRKLKHHEQ KLLKKVDFLE WKQDQGHRDT QVMRTYHIQN REDYHKYNRI CGDIRRLANK LSLLPPTDPF RRKHEQLLLD KLYAMGVLT TKSKISDLEN KVTVSAICRR RLPVIMHRLK MAETIQDAVK FIEQGHVRVG PNLINDPAYL VTRNMEDYVT W VDNSKIKK TLLRYRNQID DFDFS UniProtKB: U3 small nucleolar ribonucleoprotein protein IMP3 |
+Macromolecule #27: U3 small nucleolar ribonucleoprotein protein IMP4
| Macromolecule | Name: U3 small nucleolar ribonucleoprotein protein IMP4 / type: protein_or_peptide / ID: 27 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 33.536168 KDa |
| Sequence | String: MLRRQARERR EYLYRKAQEL QDSQLQQKRQ IIKQALAQGK PLPKELAEDE SLQKDFRYDQ SLKESEEADD LQVDDEYAAT SGIMDPRII VTTSRDPSTR LSQFAKEIKL LFPNAVRLNR GNYVMPNLVD ACKKSGTTDL VVLHEHRGVP TSLTISHFPH G PTAQFSLH ...String: MLRRQARERR EYLYRKAQEL QDSQLQQKRQ IIKQALAQGK PLPKELAEDE SLQKDFRYDQ SLKESEEADD LQVDDEYAAT SGIMDPRII VTTSRDPSTR LSQFAKEIKL LFPNAVRLNR GNYVMPNLVD ACKKSGTTDL VVLHEHRGVP TSLTISHFPH G PTAQFSLH NVVMRHDIIN AGNQSEVNPH LIFDNFTTAL GKRVVCILKH LFNAGPKKDS ERVITFANRG DFISVRQHVY VR TREGVEI AEVGPRFEMR LFELRLGTLE NKDADVEWQL RRFIRTANKK DYL UniProtKB: U3 small nucleolar ribonucleoprotein protein IMP4 |
+Macromolecule #28: Mpp10,U3 small nucleolar RNA-associated protein MPP10
| Macromolecule | Name: Mpp10,U3 small nucleolar RNA-associated protein MPP10 / type: protein_or_peptide / ID: 28 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 54.936984 KDa |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) ...String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)ELQKAH SEISELYANL VYKL DVLSS VHFVPKPAST SLEIRVETPT ISMEDAQPLY MSNASSLAPQ EIYNVGKAEK DGEIRLKNGV AMSKEELTRE DKNRL RRAL KRKRSKANLP NVNKRSKRND VVDTLSKAKN ITVINQKGEK KDVSGKTKKS RSGPDSTNIK L UniProtKB: U3 small nucleolar RNA-associated protein MPP10 |
+Macromolecule #29: Pre-rRNA-processing protein PNO1
| Macromolecule | Name: Pre-rRNA-processing protein PNO1 / type: protein_or_peptide / ID: 29 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 30.380623 KDa |
| Sequence | String: MVAPTALKKA TVTPVSGQDG GSSRIIGINN TESIDEDDDD DVLLDDSDNN TAKEEVEGEE GSRKTHESKT VVVDDQGKPR FTSASKTQG NKIKFESRKI MVPPHRMTPL RNSWTKIYPP LVEHLKLQVR MNLKTKSVEL RTNPKFTTDP GALQKGADFI K AFTLGFDL ...String: MVAPTALKKA TVTPVSGQDG GSSRIIGINN TESIDEDDDD DVLLDDSDNN TAKEEVEGEE GSRKTHESKT VVVDDQGKPR FTSASKTQG NKIKFESRKI MVPPHRMTPL RNSWTKIYPP LVEHLKLQVR MNLKTKSVEL RTNPKFTTDP GALQKGADFI K AFTLGFDL DDSIALLRLD DLYIETFEVK DVKTLTGDHL SRAIGRIAGK DGKTKFAIEN ATRTRIVLAD SKIHILGGFT HI RMARESV VSLILGSPPG KVYGNLRTVA SRLKERY UniProtKB: Pre-rRNA-processing protein PNO1 |
+Macromolecule #30: RNA 3'-terminal phosphate cyclase-like protein
| Macromolecule | Name: RNA 3'-terminal phosphate cyclase-like protein / type: protein_or_peptide / ID: 30 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 40.220559 KDa |
| Sequence | String: MSSSAPKYTT FQGSQNFRLR IVLATLSGKP IKIEKIRSGD LNPGLKDYEV SFLRLIESVT NGSVIEISYT GTTVIYRPGI IVGGASTHI CPSSKPVGYF VEPMLYLAPF SKKKFSILFK GITASHNDAG IEAIKWGLMP VMEKFGVREC ALHTLKRGSP P LGGGEVHL ...String: MSSSAPKYTT FQGSQNFRLR IVLATLSGKP IKIEKIRSGD LNPGLKDYEV SFLRLIESVT NGSVIEISYT GTTVIYRPGI IVGGASTHI CPSSKPVGYF VEPMLYLAPF SKKKFSILFK GITASHNDAG IEAIKWGLMP VMEKFGVREC ALHTLKRGSP P LGGGEVHL VVDSLIAQPI TMHEIDRPII SSITGVAYST RVSPSLVNRM IDGAKKVLKN LQCEVNITAD VWRGENSGKS PG WGITLVA QSKQKGWSYF AEDIGDAGSI PEELGEKVAC QLLEEISKSA AVGRNQLPLA IVYMVIGKED IGRLRINKEQ IDE RFIILL RDIKKIFNTE VFLKPVDEAD NEDMIATIKG IGFTNTSKKI A UniProtKB: rRNA processing protein RCL1 |
+Macromolecule #31: Sof1
| Macromolecule | Name: Sof1 / type: protein_or_peptide / ID: 31 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 41.634328 KDa |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) ...String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) |
+Macromolecule #33: 40S ribosomal protein S1-A
| Macromolecule | Name: 40S ribosomal protein S1-A / type: protein_or_peptide / ID: 33 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 28.798467 KDa |
| Sequence | String: MAVGKNKRLS KGKKGQKKRV VDPFTRKEWF DIKAPSTFEN RNVGKTLVNK STGLKSASDA LKGRVVEVCL ADLQGSEDHS FRKIKLRVD EVQGKNLLTN FHGMDFTTDK LRSMVRKWQT LIEANVTVKT SDDYVLRIFA IAFTRKQANQ VKRHSYAQSS H IRAIRKVI ...String: MAVGKNKRLS KGKKGQKKRV VDPFTRKEWF DIKAPSTFEN RNVGKTLVNK STGLKSASDA LKGRVVEVCL ADLQGSEDHS FRKIKLRVD EVQGKNLLTN FHGMDFTTDK LRSMVRKWQT LIEANVTVKT SDDYVLRIFA IAFTRKQANQ VKRHSYAQSS H IRAIRKVI SEILTKEVQG STLAQLTSKL IPEVINKEIE NATKDIFPLQ NIHVRKVKLL KQPKFDVGAL MALHGEGSGE EK GKKVTGF KDEVLETV UniProtKB: Small ribosomal subunit protein eS1A |
+Macromolecule #34: 40S ribosomal protein S4-A
| Macromolecule | Name: 40S ribosomal protein S4-A / type: protein_or_peptide / ID: 34 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 29.46933 KDa |
| Sequence | String: MARGPKKHLK RLAAPHHWLL DKLSGCYAPR PSAGPHKLRE SLPLIVFLRN RLKYALNGRE VKAILMQRHV KVDGKVRTDT TYPAGFMDV ITLDATNENF RLVYDVKGRF AVHRITDEEA SYKLGKVKKV QLGKKGVPYV VTHDGRTIRY PDPNIKVNDT V KIDLASGK ...String: MARGPKKHLK RLAAPHHWLL DKLSGCYAPR PSAGPHKLRE SLPLIVFLRN RLKYALNGRE VKAILMQRHV KVDGKVRTDT TYPAGFMDV ITLDATNENF RLVYDVKGRF AVHRITDEEA SYKLGKVKKV QLGKKGVPYV VTHDGRTIRY PDPNIKVNDT V KIDLASGK ITDFIKFDAG KLVYVTGGRN LGRIGTIVHK ERHDGGFDLV HIKDSLDNTF VTRLNNVFVI GEQGKPYISL PK GKGIKLS IAEERDRRRA QQGL UniProtKB: Small ribosomal subunit protein eS4A |
+Macromolecule #35: 40S ribosomal protein S5
| Macromolecule | Name: 40S ribosomal protein S5 / type: protein_or_peptide / ID: 35 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 25.0726 KDa |
| Sequence | String: MSDTEAPVEV QEDFEVVEEF TPVVLATPIP EEVQQAQTEI KLFNKWSFEE VEVKDASLVD YVQVRQPIFV AHTAGRYANK RFRKAQCPI IERLTNSLMM NGRNNGKKLK AVRIIKHTLD IINVLTDQNP IQVVVDAITN TGPREDTTRV GGGGAARRQA V DVSPLRRV ...String: MSDTEAPVEV QEDFEVVEEF TPVVLATPIP EEVQQAQTEI KLFNKWSFEE VEVKDASLVD YVQVRQPIFV AHTAGRYANK RFRKAQCPI IERLTNSLMM NGRNNGKKLK AVRIIKHTLD IINVLTDQNP IQVVVDAITN TGPREDTTRV GGGGAARRQA V DVSPLRRV NQAIALLTIG AREAAFRNIK TIAETLAEEL INAAKGSSTS YAIKKKDELE RVAKSNR UniProtKB: Small ribosomal subunit protein uS7 |
+Macromolecule #36: 40S ribosomal protein S7-A
| Macromolecule | Name: 40S ribosomal protein S7-A / type: protein_or_peptide / ID: 36 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 21.658209 KDa |
| Sequence | String: MSAPQAKILS QAPTELELQV AQAFVELENS SPELKAELRP LQFKSIREID VAGGKKALAI FVPVPSLAGF HKVQTKLTRE LEKKFQDRH VIFLAERRIL PKPSRTSRQV QKRPRSRTLT AVHDKILEDL VFPTEIVGKR VRYLVGGNKI QKVLLDSKDV Q QIDYKLES FQAVYNKLTG KQIVFEIPSE TH UniProtKB: Small ribosomal subunit protein eS7A |
+Macromolecule #37: 40S ribosomal protein S8-A
| Macromolecule | Name: 40S ribosomal protein S8-A / type: protein_or_peptide / ID: 37 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 22.537803 KDa |
| Sequence | String: MGISRDSRHK RSATGAKRAQ FRKKRKFELG RQPANTKIGA KRIHSVRTRG GNKKYRALRI ETGNFSWASE GISKKTRIAG VVYHPSNNE LVRTNTLTKA AIVQIDATPF RQWFEAHYGQ TLGKKKNVKE EETVAKSKNA ERKWAARAAS AKIESSVESQ F SAGRLYAC ...String: MGISRDSRHK RSATGAKRAQ FRKKRKFELG RQPANTKIGA KRIHSVRTRG GNKKYRALRI ETGNFSWASE GISKKTRIAG VVYHPSNNE LVRTNTLTKA AIVQIDATPF RQWFEAHYGQ TLGKKKNVKE EETVAKSKNA ERKWAARAAS AKIESSVESQ F SAGRLYAC ISSRPGQSGR CDGYILEGEE LAFYLRRLTA KK UniProtKB: Small ribosomal subunit protein eS8A |
+Macromolecule #38: 40S ribosomal protein S9-A
| Macromolecule | Name: 40S ribosomal protein S9-A / type: protein_or_peptide / ID: 38 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 22.487893 KDa |
| Sequence | String: MPRAPRTYSK TYSTPKRPYE SSRLDAELKL AGEFGLKNKK EIYRISFQLS KIRRAARDLL TRDEKDPKRL FEGNALIRRL VRVGVLSED KKKLDYVLAL KVEDFLERRL QTQVYKLGLA KSVHHARVLI TQRHIAVGKQ IVNIPSFMVR LDSEKHIDFA P TSPFGGAR ...String: MPRAPRTYSK TYSTPKRPYE SSRLDAELKL AGEFGLKNKK EIYRISFQLS KIRRAARDLL TRDEKDPKRL FEGNALIRRL VRVGVLSED KKKLDYVLAL KVEDFLERRL QTQVYKLGLA KSVHHARVLI TQRHIAVGKQ IVNIPSFMVR LDSEKHIDFA P TSPFGGAR PGRVARRNAA RKAEASGEAA DEADEADEE UniProtKB: Small ribosomal subunit protein uS4A |
+Macromolecule #39: 40S ribosomal protein S11-A
| Macromolecule | Name: 40S ribosomal protein S11-A / type: protein_or_peptide / ID: 39 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 17.785934 KDa |
| Sequence | String: MSTELTVQSE RAFQKQPHIF NNPKVKTSKR TKRWYKNAGL GFKTPKTAIE GSYIDKKCPF TGLVSIRGKI LTGTVVSTKM HRTIVIRRA YLHYIPKYNR YEKRHKNVPV HVSPAFRVQV GDIVTVGQCR PISKTVRFNV VKVSAAAGKA NKQFAKF UniProtKB: Small ribosomal subunit protein uS17A |
+Macromolecule #40: 40S ribosomal protein S13
| Macromolecule | Name: 40S ribosomal protein S13 / type: protein_or_peptide / ID: 40 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 17.059945 KDa |
| Sequence | String: MGRMHSAGKG ISSSAIPYSR NAPAWFKLSS ESVIEQIVKY ARKGLTPSQI GVLLRDAHGV TQARVITGNK IMRILKSNGL APEIPEDLY YLIKKAVSVR KHLERNRKDK DAKFRLILIE SRIHRLARYY RTVAVLPPNW KYESATASAL VN UniProtKB: Small ribosomal subunit protein uS15 |
+Macromolecule #41: 40S ribosomal protein S14-A
| Macromolecule | Name: 40S ribosomal protein S14-A / type: protein_or_peptide / ID: 41 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 14.562655 KDa |
| Sequence | String: MSNVVQARDN SQVFGVARIY ASFNDTFVHV TDLSGKETIA RVTGGMKVKA DRDESSPYAA MLAAQDVAAK CKEVGITAVH VKIRATGGT RTKTPGPGGQ AALRALARSG LRIGRIEDVT PVPSDSTRKK GGRRGRRL UniProtKB: Small ribosomal subunit protein uS11A |
+Macromolecule #42: 40S ribosomal protein S16-A
| Macromolecule | Name: 40S ribosomal protein S16-A / type: protein_or_peptide / ID: 42 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 15.87749 KDa |
| Sequence | String: MSAVPSVQTF GKKKSATAVA HVKAGKGLIK VNGSPITLVE PEILRFKVYE PLLLVGLDKF SNIDIRVRVT GGGHVSQVYA IRQAIAKGL VAYHQKYVDE QSKNELKKAF TSYDRTLLIA DSRRPEPKKF GGKGARSRFQ KSYR UniProtKB: Small ribosomal subunit protein uS9A |
+Macromolecule #43: 40S ribosomal protein S22-A
| Macromolecule | Name: 40S ribosomal protein S22-A / type: protein_or_peptide / ID: 43 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 14.650062 KDa |
| Sequence | String: MTRSSVLADA LNAINNAEKT GKRQVLIRPS SKVIIKFLQV MQKHGYIGEF EYIDDHRSGK IVVQLNGRLN KCGVISPRFN VKIGDIEKW TANLLPARQF GYVILTTSAG IMDHEEARRK HVSGKILGFV Y UniProtKB: Small ribosomal subunit protein uS8A |
+Macromolecule #44: 40S ribosomal protein S23-A
| Macromolecule | Name: 40S ribosomal protein S23-A / type: protein_or_peptide / ID: 44 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 16.073896 KDa |
| Sequence | String: MGKGKPRGLN SARKLRVHRR NNRWAENNYK KRLLGTAFKS SPFGGSSHAK GIVLEKLGIE SKQPNSAIRK CVRVQLIKNG KKVTAFVPN DGCLNFVDEN DEVLLAGFGR KGKAKGDIPG VRFKVVKVSG VSLLALWKEK KEKPRS UniProtKB: Small ribosomal subunit protein uS12A |
+Macromolecule #45: 40S ribosomal protein S24-A
| Macromolecule | Name: 40S ribosomal protein S24-A / type: protein_or_peptide / ID: 45 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 15.362848 KDa |
| Sequence | String: MSDAVTIRTR KVISNPLLAR KQFVVDVLHP NRANVSKDEL REKLAEVYKA EKDAVSVFGF RTQFGGGKSV GFGLVYNSVA EAKKFEPTY RLVRYGLAEK VEKASRQQRK QKKNRDKKIF GTGKRLAKKV ARRNAD UniProtKB: Small ribosomal subunit protein eS24A |
+Macromolecule #46: 40S ribosomal protein S27-A
| Macromolecule | Name: 40S ribosomal protein S27-A / type: protein_or_peptide / ID: 46 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 8.893391 KDa |
| Sequence | String: MVLVQDLLHP TAASEARKHK LKTLVQGPRS YFLDVKCPGC LNITTVFSHA QTAVTCESCS TILCTPTGGK AKLSEGTSFR RK UniProtKB: Small ribosomal subunit protein eS27A |
+Macromolecule #47: 40S ribosomal protein S28-A
| Macromolecule | Name: 40S ribosomal protein S28-A / type: protein_or_peptide / ID: 47 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 7.605847 KDa |
| Sequence | String: MDNKTPVTLA KVIKVLGRTG SRGGVTQVRV EFLEDTSRTI VRNVKGPVRE NDILVLMESE REARRLR UniProtKB: Small ribosomal subunit protein eS28A |
+Macromolecule #48: 40S ribosomal protein S30-A
| Macromolecule | Name: 40S ribosomal protein S30-A / type: protein_or_peptide / ID: 48 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 7.137541 KDa |
| Sequence | String: MAKVHGSLAR AGKVKSQTPK VEKTEKPKKP KGRAYKRLLY TRRFVNVTLV NGKRRMNPGP SVQ UniProtKB: Small ribosomal subunit protein eS30A |
+Macromolecule #49: Utp7
| Macromolecule | Name: Utp7 / type: protein_or_peptide / ID: 49 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 47.166211 KDa |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) ...String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) |
+Macromolecule #50: Utp11
| Macromolecule | Name: Utp11 / type: protein_or_peptide / ID: 50 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 21.294152 KDa |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) ...String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) |
+Macromolecule #51: rRNA-processing protein FCF1
| Macromolecule | Name: rRNA-processing protein FCF1 / type: protein_or_peptide / ID: 51 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 21.650729 KDa |
| Sequence | String: MGKAKKTRKF GLVKRTLNTK KDQRLKKNQE NIKTKEDPEL TRNIPQVSSA LFFQYNQAIK PPYQVLIDTN FINFSIQKKV DIVRGMMDC LLAKCNPLIT DCVMAELEKL GPKYRIALKL ARDPRIKRLS CSHKGTYADD CLVHRVLQHK CYIVATNDAG L KQRIRKIP GIPLMSVGGH AYVIEKLPDV F UniProtKB: rRNA-processing protein FCF1 |
+Macromolecule #52: Ribosome biogenesis protein UTP30
| Macromolecule | Name: Ribosome biogenesis protein UTP30 / type: protein_or_peptide / ID: 52 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 31.668939 KDa |
| Sequence | String: MVESNDIIKS GLAEKALKAL ILQCEENPSL KNDKDIHIII NTGKKMGINR DNIPRIIPLT KYKLFKPRDL NILLITKDPS ALYRETLTK DEHTSELFKE IISVKNLRRR FKGSKLTQLY KDFDLVVADY RVHHLLPEVL GSRFYHGSKK LPYMIRMSKE V KLKRQQMV ...String: MVESNDIIKS GLAEKALKAL ILQCEENPSL KNDKDIHIII NTGKKMGINR DNIPRIIPLT KYKLFKPRDL NILLITKDPS ALYRETLTK DEHTSELFKE IISVKNLRRR FKGSKLTQLY KDFDLVVADY RVHHLLPEVL GSRFYHGSKK LPYMIRMSKE V KLKRQQMV EKCDPIYVRA QLRSICKNTS YIPNNDNCLS VRVGYIQKHS IPEILQNIQD TINFLTDKSK RPQGGVIKGG II SIFVKTS NSTSLPIYQF SEARENQKNE DLSDIKL UniProtKB: Ribosome biogenesis protein UTP30 |
+Macromolecule #53: Helical domain protein
| Macromolecule | Name: Helical domain protein / type: protein_or_peptide / ID: 53 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 137.462422 KDa |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) ...String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) |
+Macromolecule #54: Helical domain protein
| Macromolecule | Name: Helical domain protein / type: protein_or_peptide / ID: 54 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 84.015695 KDa |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) ...String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) |
+Macromolecule #55: Unassigned helices
| Macromolecule | Name: Unassigned helices / type: protein_or_peptide / ID: 55 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 87.930383 KDa |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) ...String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 Component:
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 5 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 15 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 101.325 kPa | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV / Details: blot for 2 seconds before plunging. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: OTHER / Digitization - Dimensions - Width: 4000 pixel / Digitization - Dimensions - Height: 4000 pixel / Number grids imaged: 1 / Number real images: 1102 / Average exposure time: 1.6 sec. / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 79545 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 45000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
+ About Yorodumi
About Yorodumi
-News
-Feb 9, 2022. New format data for meta-information of EMDB entries
New format data for meta-information of EMDB entries
- Version 3 of the EMDB header file is now the official format.
- The previous official version 1.9 will be removed from the archive.
Related info.: EMDB header
EMDB header
External links: wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
-Aug 12, 2020. Covid-19 info
Covid-19 info

URL: https://pdbj.org/emnavi/covid19.php
New page: Covid-19 featured information page in EM Navigator.
Related info.: Covid-19 info /
Covid-19 info /  Mar 5, 2020. Novel coronavirus structure data
Mar 5, 2020. Novel coronavirus structure data
+Mar 5, 2020. Novel coronavirus structure data
Novel coronavirus structure data
- International Committee on Taxonomy of Viruses (ICTV) defined the short name of the 2019 coronavirus as "SARS-CoV-2".
- In the structure databanks used in Yorodumi, some data are registered as the other names, "COVID-19 virus" and "2019-nCoV". Here are the details of the virus and the list of structure data.
Related info.: Yorodumi Speices /
Yorodumi Speices /  Aug 12, 2020. Covid-19 info
Aug 12, 2020. Covid-19 info
External links: COVID-19 featured content - PDBj /
COVID-19 featured content - PDBj /  Molecule of the Month (242):Coronavirus Proteases
Molecule of the Month (242):Coronavirus Proteases
+Jan 31, 2019. EMDB accession codes are about to change! (news from PDBe EMDB page)
EMDB accession codes are about to change! (news from PDBe EMDB page)
- The allocation of 4 digits for EMDB accession codes will soon come to an end. Whilst these codes will remain in use, new EMDB accession codes will include an additional digit and will expand incrementally as the available range of codes is exhausted. The current 4-digit format prefixed with “EMD-” (i.e. EMD-XXXX) will advance to a 5-digit format (i.e. EMD-XXXXX), and so on. It is currently estimated that the 4-digit codes will be depleted around Spring 2019, at which point the 5-digit format will come into force.
- The EM Navigator/Yorodumi systems omit the EMD- prefix.
Related info.: Q: What is EMD? /
Q: What is EMD? /  ID/Accession-code notation in Yorodumi/EM Navigator
ID/Accession-code notation in Yorodumi/EM Navigator
External links: EMDB Accession Codes are Changing Soon! /
EMDB Accession Codes are Changing Soon! /  Contact to PDBj
Contact to PDBj
+Jul 12, 2017. Major update of PDB
Major update of PDB
- wwPDB released updated PDB data conforming to the new PDBx/mmCIF dictionary.
- This is a major update changing the version number from 4 to 5, and with Remediation, in which all the entries are updated.
- In this update, many items about electron microscopy experimental information are reorganized (e.g. em_software).
- Now, EM Navigator and Yorodumi are based on the updated data.
External links: wwPDB Remediation /
wwPDB Remediation /  Enriched Model Files Conforming to OneDep Data Standards Now Available in the PDB FTP Archive
Enriched Model Files Conforming to OneDep Data Standards Now Available in the PDB FTP Archive
-Yorodumi
Thousand views of thousand structures
- Yorodumi is a browser for structure data from EMDB, PDB, SASBDB, etc.
- This page is also the successor to EM Navigator detail page, and also detail information page/front-end page for Omokage search.
- The word "yorodu" (or yorozu) is an old Japanese word meaning "ten thousand". "mi" (miru) is to see.
Related info.: EMDB /
EMDB /  PDB /
PDB /  SASBDB /
SASBDB /  Comparison of 3 databanks /
Comparison of 3 databanks /  Yorodumi Search /
Yorodumi Search /  Aug 31, 2016. New EM Navigator & Yorodumi /
Aug 31, 2016. New EM Navigator & Yorodumi /  Yorodumi Papers /
Yorodumi Papers /  Jmol/JSmol /
Jmol/JSmol /  Function and homology information /
Function and homology information /  Changes in new EM Navigator and Yorodumi
Changes in new EM Navigator and Yorodumi
 Movie
Movie Controller
Controller



























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)