+ Open data
Open data
 Loading...
Loading...
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8859 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | The complete structure of the small subunit processome | ||||||||||||||||||
 Map data Map data | small subunit processome, overall map 1 | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | Ribosome assembly / RIBOSOME | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationtRNA wobble cytosine modification / tRNA cytidine N4-acetyltransferase activity / rRNA acetylation involved in maturation of SSU-rRNA / 18S rRNA cytidine N-acetyltransferase activity / regulation of ribosomal protein gene transcription by RNA polymerase II / tRNA acetylation / box H/ACA snoRNA binding / RNA fragment catabolic process / rRNA small subunit pseudouridine methyltransferase Nep1 / CURI complex ...tRNA wobble cytosine modification / tRNA cytidine N4-acetyltransferase activity / rRNA acetylation involved in maturation of SSU-rRNA / 18S rRNA cytidine N-acetyltransferase activity / regulation of ribosomal protein gene transcription by RNA polymerase II / tRNA acetylation / box H/ACA snoRNA binding / RNA fragment catabolic process / rRNA small subunit pseudouridine methyltransferase Nep1 / CURI complex / UTP-C complex / rRNA 2'-O-methylation / Noc4p-Nop14p complex / endonucleolytic cleavage in ITS1 upstream of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / t-UTP complex / Pwp2p-containing subcomplex of 90S preribosome / nuclear microtubule / Mpp10 complex / rRNA (pseudouridine) methyltransferase activity / box C/D sno(s)RNA binding / snoRNA guided rRNA 2'-O-methylation / rRNA modification / histone H2AQ104 methyltransferase activity / septum digestion after cytokinesis / regulation of rRNA processing / tRNA re-export from nucleus / snRNA binding / box C/D sno(s)RNA 3'-end processing / RNA folding chaperone / rRNA methyltransferase activity / rDNA heterochromatin / endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / regulation of transcription by RNA polymerase I / endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / box C/D methylation guide snoRNP complex / U4/U6 snRNP / positive regulation of rRNA processing / tRNA export from nucleus / single-stranded telomeric DNA binding / rRNA primary transcript binding / sno(s)RNA-containing ribonucleoprotein complex / O-methyltransferase activity / small nuclear ribonucleoprotein complex / rRNA base methylation / U4 snRNA binding / SUMOylation of RNA binding proteins / protein localization to nucleolus / rRNA methylation / mTORC1-mediated signalling / mRNA modification / 90S preribosome assembly / U4 snRNP / poly(U) RNA binding / U3 snoRNA binding / positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay / Formation of the ternary complex, and subsequently, the 43S complex / Translation initiation complex formation / Ribosomal scanning and start codon recognition / poly(A)+ mRNA export from nucleus / snoRNA binding / precatalytic spliceosome / preribosome, small subunit precursor / Major pathway of rRNA processing in the nucleolus and cytosol / establishment of cell polarity / spliceosomal complex assembly / positive regulation of transcription by RNA polymerase I / SRP-dependent cotranslational protein targeting to membrane / GTP hydrolysis and joining of the 60S ribosomal subunit / nucleolar large rRNA transcription by RNA polymerase I / Formation of a pool of free 40S subunits / Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) / Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) / L13a-mediated translational silencing of Ceruloplasmin expression / 90S preribosome / proteasome assembly / RNA processing / ribosomal subunit export from nucleus / U4/U6 x U5 tri-snRNP complex / regulation of translational fidelity / endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / ribosomal small subunit export from nucleus / RNA endonuclease activity / Transferases; Acyltransferases; Transferring groups other than aminoacyl groups / nuclear periphery / Transferases; Transferring one-carbon groups; Methyltransferases / ribosome assembly / maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / spliceosomal complex / maturation of SSU-rRNA / translational initiation / small-subunit processome / enzyme activator activity / mRNA splicing, via spliceosome / maintenance of translational fidelity / rRNA processing / unfolded protein binding / peroxisome / ribosome biogenesis / ribosomal small subunit biogenesis Similarity search - Function Regulator of rDNA transcription 14 / Regular of rDNA transcription protein 14 / : / NUC153 / Nucleolar protein 10/Enp2 / : / : / NUC153 domain / Nucleolar protein 10-like, second domain / Nucleolar protein 10-like, N-terminal domain ...Regulator of rDNA transcription 14 / Regular of rDNA transcription protein 14 / : / NUC153 / Nucleolar protein 10/Enp2 / : / : / NUC153 domain / Nucleolar protein 10-like, second domain / Nucleolar protein 10-like, N-terminal domain / rRNA biogenesis protein Rrp5 / : / : / Ribosomal RNA assembly KRR1 / : / : / : / KRR1 small subunit processome component, second KH domain / U3 small nucleolar RNA-associated protein 8 / : / Utp8, N-terminal beta propeller / Utp8, C-terminal / Possible tRNA binding domain / RNA cytidine acetyltransferase NAT10 / Possible tRNA binding domain / Helicase domain / tRNA(Met) cytidine acetyltransferase TmcA, N-terminal / TmcA/NAT10/Kre33 / RNA cytidine acetyltransferase NAT10/TcmA, helicase domain / tRNA(Met) cytidine acetyltransferase TmcA, N-terminal / GNAT acetyltransferase 2 / : / U3 snoRNA associated / U3 snoRNA associated / : / Sof1-like protein / : / Sof1-like domain / WD repeat-containing protein 75 second beta-propeller / NOL6/Upt22 / BING4, C-terminal domain / Nucleolar complex protein 4 / Nrap protein domain 1 / Nrap protein, domain 2 / Nrap protein, domain 3 / Nrap protein, domain 4 / Nrap protein, domain 5 / Nrap protein, domain 6 / WD repeat-containing protein WDR46/Utp7 / Nrap protein domain 1 / BING4CT (NUC141) domain / Nrap protein PAP/OAS-like domain / Nrap protein domain 3 / Nrap protein nucleotidyltransferase domain 4 / Nrap protein PAP/OAS1-like domain 5 / Nrap protein domain 6 / BING4CT (NUC141) domain / U3 small nucleolar RNA-associated protein 6 / Ribosomal RNA-processing protein 7, C-terminal domain / Ribosomal RNA-processing protein 7 / Rrp7, RRM-like N-terminal domain / : / U3 small nucleolar RNA-associated protein 6 / Ribosomal RNA-processing protein 7 (RRP7) C-terminal domain / Rrp7 RRM-like N-terminal domain / U3 small nucleolar RNA-associated protein 4 / Small-subunit processome, Utp11 / Nucleolar protein 14 / Fcf2 pre-rRNA processing, C-terminal / Fcf2/DNTTIP2 / : / Utp11 protein / Nop14-like family / Fcf2 pre-rRNA processing / : / U3 small nucleolar RNA-associated protein 15, C-terminal / UTP15 C terminal / rRNA-processing protein Fcf1, PIN domain / WDR3 second beta-propeller domain / WDR3 first beta-propeller domain / Small-subunit processome, Utp14 / U3 small nucleolar RNA-associated protein 18 / Utp14 protein / U3 small nucleolar RNA-associated protein 13, C-terminal / Periodic tryptophan protein 2 / Utp13 specific WD40 associated domain / BP28, C-terminal domain / Nucleolar protein 58/56, N-terminal / U3 small nucleolar RNA-associated protein 10, N-terminal / U3 small nucleolar RNA-associated protein 10 / : / BP28CT (NUC211) domain / NOP5NT (NUC127) domain / U3 small nucleolar RNA-associated protein 10 / HEATR1-like, HEAT repeats / BP28CT (NUC211) domain / rRNA-processing protein Fcf1/Utp23 / Sas10 C-terminal domain / : / Fcf1 Similarity search - Domain/homology Rps5p / KRR1 small subunit processome component / Regulator of rDNA transcription 14 / U3 small nucleolar RNA-associated protein 11 / Regulator of rDNA transcription 14 / Small ribosomal subunit protein uS4A / Small ribosomal subunit protein uS15 / Small ribosomal subunit protein uS11A / Small ribosomal subunit protein uS8A / Small ribosomal subunit protein eS24A ...Rps5p / KRR1 small subunit processome component / Regulator of rDNA transcription 14 / U3 small nucleolar RNA-associated protein 11 / Regulator of rDNA transcription 14 / Small ribosomal subunit protein uS4A / Small ribosomal subunit protein uS15 / Small ribosomal subunit protein uS11A / Small ribosomal subunit protein uS8A / Small ribosomal subunit protein eS24A / Small ribosomal subunit protein eS4A / Small ribosomal subunit protein eS6A / Small ribosomal subunit protein eS8A / Small ribosomal subunit protein uS17A / Small ribosomal subunit protein uS9A / Small ribosomal subunit protein uS13A / Small ribosomal subunit protein uS12A / rRNA 2'-O-methyltransferase fibrillarin / Ribosomal RNA-processing protein 7 / Periodic tryptophan protein 2 / Small ribosomal subunit protein eS7A / U3 small nucleolar ribonucleoprotein protein IMP3 / Protein SOF1 / Ribosome biogenesis protein UTP30 / Essential nuclear protein 1 / U3 small nucleolar RNA-associated protein 9 / 13 kDa ribonucleoprotein-associated protein / U3 small nucleolar RNA-associated protein 7 / U3 small nucleolar ribonucleoprotein protein LCP5 / U3 small nucleolar RNA-associated protein 18 / Protein FAF1 / U3 small nucleolar RNA-associated protein 10 / U3 small nucleolar RNA-associated protein MPP10 / Ribosome biogenesis protein ENP2 / U3 small nucleolar RNA-associated protein 22 / U3 small nucleolar RNA-associated protein 8 / RNA cytidine acetyltransferase / U3 small nucleolar ribonucleoprotein protein IMP4 / U3 small nucleolar RNA-associated protein 6 / NET1-associated nuclear protein 1 / U3 small nucleolar RNA-associated protein 5 / U3 small nucleolar RNA-associated protein 15 / U3 small nucleolar RNA-associated protein 14 / rRNA biogenesis protein RRP5 / rRNA-processing protein FCF1 / U3 small nucleolar RNA-associated protein 13 / U3 small nucleolar RNA-associated protein 21 / Ribosomal RNA small subunit methyltransferase NEP1 / Ribosomal RNA-processing protein 9 / Nucleolar complex protein 4 / U3 small nucleolar RNA-associated protein 4 / rRNA processing protein RCL1 / U3 small nucleolar RNA-associated protein 16 / Ribosome biogenesis protein BMS1 / rRNA-processing protein FCF2 / Something about silencing protein 10 / U3 small nucleolar RNA-associated protein 12 / Nucleolar protein 56 / Nucleolar protein 58 / Small ribosomal subunit protein eS28A / Nucleolar complex protein 14 / Pre-rRNA-processing protein PNO1 Similarity search - Component | ||||||||||||||||||
| Biological species |  | ||||||||||||||||||
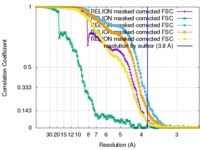
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | ||||||||||||||||||
 Authors Authors | Barandun J / Chaker-Margot M | ||||||||||||||||||
| Funding support |  United States, European Union, United States, European Union,  Switzerland, Switzerland,  Canada, Canada,  France, 5 items France, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2017 Journal: Nat Struct Mol Biol / Year: 2017Title: The complete structure of the small-subunit processome. Authors: Jonas Barandun / Malik Chaker-Margot / Mirjam Hunziker / Kelly R Molloy / Brian T Chait / Sebastian Klinge /  Abstract: The small-subunit processome represents the earliest stable precursor of the eukaryotic small ribosomal subunit. Here we present the cryo-EM structure of the Saccharomyces cerevisiae small-subunit ...The small-subunit processome represents the earliest stable precursor of the eukaryotic small ribosomal subunit. Here we present the cryo-EM structure of the Saccharomyces cerevisiae small-subunit processome at an overall resolution of 3.8 Å, which provides an essentially complete near-atomic model of this assembly. In this nucleolar superstructure, 51 ribosome-assembly factors and two RNAs encapsulate the 18S rRNA precursor and 15 ribosomal proteins in a state that precedes pre-rRNA cleavage at site A1. Extended flexible proteins are employed to connect distant sites in this particle. Molecular mimicry and steric hindrance, as well as protein- and RNA-mediated RNA remodeling, are used in a concerted fashion to prevent the premature formation of the central pseudoknot and its surrounding elements within the small ribosomal subunit. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8859.map.gz emd_8859.map.gz | 223 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8859-v30.xml emd-8859-v30.xml emd-8859.xml emd-8859.xml | 131.5 KB 131.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_8859_fsc_1.xml emd_8859_fsc_1.xml emd_8859_fsc_2.xml emd_8859_fsc_2.xml emd_8859_fsc_3.xml emd_8859_fsc_3.xml emd_8859_fsc_4.xml emd_8859_fsc_4.xml emd_8859_fsc_5.xml emd_8859_fsc_5.xml | 13.9 KB 13.8 KB 13.9 KB 13.9 KB 13.9 KB | Display Display Display Display Display |  FSC data file FSC data file |
| Images |  emd_8859.png emd_8859.png | 211.6 KB | ||
| Filedesc metadata |  emd-8859.cif.gz emd-8859.cif.gz | 27.5 KB | ||
| Others |  emd_8859_additional_1.map.gz emd_8859_additional_1.map.gz emd_8859_additional_2.map.gz emd_8859_additional_2.map.gz emd_8859_additional_3.map.gz emd_8859_additional_3.map.gz emd_8859_additional_4.map.gz emd_8859_additional_4.map.gz | 215.9 MB 229 MB 227.4 MB 224.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8859 http://ftp.pdbj.org/pub/emdb/structures/EMD-8859 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8859 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8859 | HTTPS FTP |
-Related structure data
| Related structure data |  5wlcMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_8859.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8859.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | small subunit processome, overall map 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.3 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
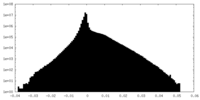
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
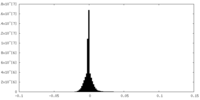
-Additional map: small subunit processome, central domain map
| File | emd_8859_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | small subunit processome, central domain map | ||||||||||||
| Projections & Slices |
| ||||||||||||
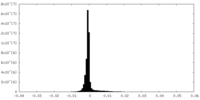
| Density Histograms |
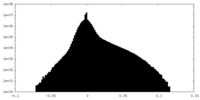
-Additional map: small subunit processome, core map
| File | emd_8859_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | small subunit processome, core map | ||||||||||||
| Projections & Slices |
| ||||||||||||
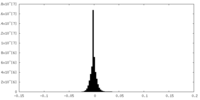
| Density Histograms |
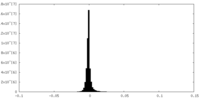
-Additional map: small subunit processome, overall map 2
| File | emd_8859_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | small subunit processome, overall map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
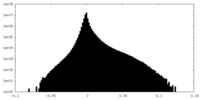
-Additional map: small subunit processome, 3' domain map
| File | emd_8859_additional_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | small subunit processome, 3' domain map | ||||||||||||
| Projections & Slices |
| ||||||||||||
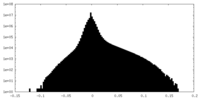
| Density Histograms |
- Sample components
Sample components
+Entire : Small subunit processome
| Entire | Name: Small subunit processome |
|---|---|
| Components |
|
+Supramolecule #1: Small subunit processome
| Supramolecule | Name: Small subunit processome / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#66 |
|---|---|
| Source (natural) | Organism:  |
+Macromolecule #1: 5' ETS
| Macromolecule | Name: 5' ETS / type: rna / ID: 1 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 225.543094 KDa |
| Sequence | String: AUGCGAAAGC AGUUGAAGAC AAGUUCGAAA AGAGUUUGGA AACGAAUUCG AGUAGGCUUG UCGUUCGUUA UGUUUUUGUA AAUGGCCUC GUCAAACGGU GGAGAGAGUC GCUAGGUGAU CGUCAGAUCU GCCUAGUCUC UAUACAGCGU GUUUAAUUGA C AUGGGUUG ...String: AUGCGAAAGC AGUUGAAGAC AAGUUCGAAA AGAGUUUGGA AACGAAUUCG AGUAGGCUUG UCGUUCGUUA UGUUUUUGUA AAUGGCCUC GUCAAACGGU GGAGAGAGUC GCUAGGUGAU CGUCAGAUCU GCCUAGUCUC UAUACAGCGU GUUUAAUUGA C AUGGGUUG AUGCGUAUUG AGAGAUACAA UUUGGGAAGA AAUUCCCAGA GUGUGUUUCU UUUGCGUUUA ACCUGAACAG UC UCAUCGU GGGCAUCUUG CGAUUCCAUU GGUGAGCAGC GAAGGAUUUG GUGGAUUACU AGCUAAUAGC AAUCUAUUUC AAA GAAUUC AAACUUGGGG GAAUGCCUUG UUGAAUAGCC GGUCGCAAGA CUGUGAUUCU UCAAGUGUAA CCUCCUCUCA AAUC AGCGA UAUCAAACGU ACCAUUCCGU GAAACACCGG GGUAUCUGUU UGGUGGAACC UGAUUAGAGG AAACUCAAAG AGUGC UAUG GUAUGGUGAC GGAGUGCGCU GGUCAAGAGU GUAAAAGCUU UUUGAACAGA GAGCAUUUCC GGCAGCAGAG AGACCU GAA AAAGCAAUUU UUCUGGAAUU UCAGCUGUUU CCAAACUCAA UAAGUAUCUU CUAGCAAGAG GGAAUAGGUG GGAAAAA AA AAAAGAGAUU UCGGUUUCUU UCUUUUUUAC UGCUUGUUGC UUCUUCUUUU AAGAUAGU GENBANK: GENBANK: U53879.1 |
+Macromolecule #2: 18S pre-rRNA
| Macromolecule | Name: 18S pre-rRNA / type: rna / ID: 2 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 582.052438 KDa |
| Sequence | String: AGAUAGUUAU CUGGUUGAUC CUGCCAGUAG UCAUAUGCUU GUCUCAAAGA UUAAGCCAUG CAUGUCUAAG UAUAAGCAAU UUAUACAGU GAAACUGCGA AUGGCUCAUU AAAUCAGUUA UCGUUUAUUU GAUAGUUCCU UUACUACAUG GUAUAACUGU G GUAAUUCU ...String: AGAUAGUUAU CUGGUUGAUC CUGCCAGUAG UCAUAUGCUU GUCUCAAAGA UUAAGCCAUG CAUGUCUAAG UAUAAGCAAU UUAUACAGU GAAACUGCGA AUGGCUCAUU AAAUCAGUUA UCGUUUAUUU GAUAGUUCCU UUACUACAUG GUAUAACUGU G GUAAUUCU AGAGCUAAUA CAUGCUUAAA AUCUCGACCC UUUGGAAGAG AUGUAUUUAU UAGAUAAAAA AUCAAUGUCU UC GGACUCU UUGAUGAUUC AUAAUAACUU UUCGAAUCGC AUGGCCUUGU GCUGGCGAUG GUUCAUUCAA AUUUCUGCCC UAU CAACUU UCGAUGGUAG GAUAGUGGCC UACCAUGGUU UCAACGGGUA ACGGGGAAUA AGGGUUCGAU UCCGGAGAGG GAGC CUGAG AAACGGCUAC CACAUCCAAG GAAGGCAGCA GGCGCGCAAA UUACCCAAUC CUAAUUCAGG GAGGUAGUGA CAAUA AAUA ACGAUACAGG GCCCAUUCGG GUCUUGUAAU UGGAAUGAGU ACAAUGUAAA UACCUUAACG AGGAACAAUU GGAGGG CAA GUCUGGUGCC AGCAGCCGCG GUAAUUCCAG CUCCAAUAGC GUAUAUUAAA GUUGUUGCAG UUAAAAAGCU CGUAGUU GA ACUUUGGGCC CGGUUGGCCG GUCCGAUUUU UUCGUGUACU GGAUUUCCAA CGGGGCCUUU CCUUCUGGCU AACCUUGA G UCCUUGUGGC UCUUGGCGAA CCAGGACUUU UACUUUGAAA AAAUUAGAGU GUUCAAAGCA GGCGUAUUGC UCGAAUAUA UUAGCAUGGA AUAAUAGAAU AGGACGUUUG GUUCUAUUUU GUUGGUUUCU AGGACCAUCG UAAUGAUUAA UAGGGACGGU CGGGGGCAU CAGUAUUCAA UUGUCAGAGG UGAAAUUCUU GGAUUUAUUG AAGACUAACU ACUGCGAAAG CAUUUGCCAA G GACGUUUU CAUUAAUCAA GAACGAAAGU UAGGGGAUCG AAGAUGAUCA GAUACCGUCG UAGUCUUAAC CAUAAACUAU GC CGACUAG GGAUCGGGUG GUGUUUUUUU AAUGACCCAC UCGGCACCUU ACGAGAAAUC AAAGUCUUUG GGUUCUGGGG GGA GUAUGG UCGCAAGGCU GAAACUUAAA GGAAUUGACG GAAGGGCACC ACCAGGAGUG GAGCCUGCGG CUUAAUUUGA CUCA ACACG GGGAAACUCA CCAGGUCCAG ACACAAUAAG GAUUGACAGA UUGAGAGCUC UUUCUUGAUU UUGUGGGUGG UGGUG CAUG GCCGUUCUUA GUUGGUGGAG UGAUUUGUCU GCUUAAUUGC GAUAACGAAC GAGACCUUAA CCUACUAAAU AGUGGU GCU AGCAUUUGCU GGUUAUCCAC UUCUUAGAGG GACUAUCGGU UUCAAGCCGA UGGAAGUUUG AGGCAAUAAC AGGUCUG UG AUGCCCUUAG ACGUUCUGGG CCGCACGCGC GCUACACUGA CGGAGCCAGC GAGUCUAACC UUGGCCGAGA GGUCUUGG U AAUCUUGUGA AACUCCGUCG UGCUGGGGAU AGAGCAUUGU AAUUAUUGCU CUUCAACGAG GAAUUCCUAG UAAGCGCAA GUCAUCAGCU UGCGUUGAUU ACGUCCCUGC CCUUUGUACA CACCGCCCGU CGCUAGUACC GAUUGAAUGG CUUAGUGAGG CCUCAGGAU CUGCUUAGAG AAGGGGGCAA CUCCAUCUCA GAGCGGAGAA UUUGGACAAA CUUGGUCAUU UAGAGGAACU A AAAGUCGU AACAAGGUUU CCGUAGGUGA ACCUGCGGAA GGAUCAUUA GENBANK: GENBANK: CP011558.1 |
+Macromolecule #3: U3 snoRNA
| Macromolecule | Name: U3 snoRNA / type: rna / ID: 3 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 106.503258 KDa |
| Sequence | String: GUCGACGUAC UUCAUAGGAU CAUUUCUAUA GGAAUCGUCA CUCUUUGACU CUUCAAAAGA GCCACUGAAU CCAACUUGGU UGAUGAGUC CCAUAACCUU UGUACCCCAG AGUGAGAAAC CGAAAUUGAA UCUAAAUUAG CUUGGUCCGC AAUCCUUAGC G GUUCGGCC ...String: GUCGACGUAC UUCAUAGGAU CAUUUCUAUA GGAAUCGUCA CUCUUUGACU CUUCAAAAGA GCCACUGAAU CCAACUUGGU UGAUGAGUC CCAUAACCUU UGUACCCCAG AGUGAGAAAC CGAAAUUGAA UCUAAAUUAG CUUGGUCCGC AAUCCUUAGC G GUUCGGCC AUCUAUAAUU UUGAAUAAAA AUUUUGCUUU GCCGUUGCAU UUGUAGUUUU UUCCUUUGGA AGUAAUUACA AU AUUUUAU GGCGCGAUGA UCUUGACCCA UCCUAUGUAC UUCUUUUUUG AAGGGAUAGG GCUCUAUGGG UGGGUACAAA UGG CAGUCU GACAAGU GENBANK: GENBANK: M26649.1 |
+Macromolecule #4: rpS18_uS13
| Macromolecule | Name: rpS18_uS13 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 16.773332 KDa |
| Sequence | String: MSLVVQEQGS FQHILRLLNT NVDGNIKIVY ALTTIKGVGR RYSNLVCKKA DVDLHKRAGE LTQEELERIV QIMQNPTHYK IPAWFLNRQ NDITDGKDYH TLANNVESKL RDDLERLKKI R(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) WGLRVRGQHT KTTGRRRA UniProtKB: Small ribosomal subunit protein uS13A, Small ribosomal subunit protein uS13A |
+Macromolecule #5: rpS4_eS4
| Macromolecule | Name: rpS4_eS4 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 29.46933 KDa |
| Sequence | String: MARGPKKHLK RLAAPHHWLL DKLSGCYAPR PSAGPHKLRE SLPLIVFLRN RLKYALNGRE VKAILMQRHV KVDGKVRTDT TYPAGFMDV ITLDATNENF RLVYDVKGRF AVHRITDEEA SYKLGKVKKV QLGKKGVPYV VTHDGRTIRY PDPNIKVNDT V KIDLASGK ...String: MARGPKKHLK RLAAPHHWLL DKLSGCYAPR PSAGPHKLRE SLPLIVFLRN RLKYALNGRE VKAILMQRHV KVDGKVRTDT TYPAGFMDV ITLDATNENF RLVYDVKGRF AVHRITDEEA SYKLGKVKKV QLGKKGVPYV VTHDGRTIRY PDPNIKVNDT V KIDLASGK ITDFIKFDAG KLVYVTGGRN LGRIGTIVHK ERHDGGFDLV HIKDSLDNTF VTRLNNVFVI GEQGKPYISL PK GKGIKLS IAEERDRRRA QQGL UniProtKB: Small ribosomal subunit protein eS4A |
+Macromolecule #6: rpS5_uS7
| Macromolecule | Name: rpS5_uS7 / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 25.0726 KDa |
| Sequence | String: MSDTEAPVEV QEDFEVVEEF TPVVLATPIP EEVQQAQTEI KLFNKWSFEE VEVKDASLVD YVQVRQPIFV AHTAGRYANK RFRKAQCPI IERLTNSLMM NGRNNGKKLK AVRIIKHTLD IINVLTDQNP IQVVVDAITN TGPREDTTRV GGGGAARRQA V DVSPLRRV ...String: MSDTEAPVEV QEDFEVVEEF TPVVLATPIP EEVQQAQTEI KLFNKWSFEE VEVKDASLVD YVQVRQPIFV AHTAGRYANK RFRKAQCPI IERLTNSLMM NGRNNGKKLK AVRIIKHTLD IINVLTDQNP IQVVVDAITN TGPREDTTRV GGGGAARRQA V DVSPLRRV NQAIALLTIG AREAAFRNIK TIAETLAEEL INAAKGSSTS YAIKKKDELE RVAKSNR UniProtKB: Rps5p |
+Macromolecule #7: rpS6_eS6
| Macromolecule | Name: rpS6_eS6 / type: protein_or_peptide / ID: 7 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 27.054486 KDa |
| Sequence | String: MKLNISYPVN GSQKTFEIDD EHRIRVFFDK RIGQEVDGEA VGDEFKGYVF KISGGNDKQG FPMKQGVLLP TRIKLLLTKN VSCYRPRRD GERKRKSVRG AIVGPDLAVL ALVIVKKGEQ ELEGLTDTTV PKRLGPKRAN NIRKFFGLSK EDDVRDFVIR R EVTKGEKT ...String: MKLNISYPVN GSQKTFEIDD EHRIRVFFDK RIGQEVDGEA VGDEFKGYVF KISGGNDKQG FPMKQGVLLP TRIKLLLTKN VSCYRPRRD GERKRKSVRG AIVGPDLAVL ALVIVKKGEQ ELEGLTDTTV PKRLGPKRAN NIRKFFGLSK EDDVRDFVIR R EVTKGEKT YTKAPKIQRL VTPQRLQRKR HQRALKVRNA QAQREAAAEY AQLLAKRLSE RKAEKAEIRK RRASSLKA UniProtKB: Small ribosomal subunit protein eS6A |
+Macromolecule #8: rpS7_eS7
| Macromolecule | Name: rpS7_eS7 / type: protein_or_peptide / ID: 8 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 21.658209 KDa |
| Sequence | String: MSAPQAKILS QAPTELELQV AQAFVELENS SPELKAELRP LQFKSIREID VAGGKKALAI FVPVPSLAGF HKVQTKLTRE LEKKFQDRH VIFLAERRIL PKPSRTSRQV QKRPRSRTLT AVHDKILEDL VFPTEIVGKR VRYLVGGNKI QKVLLDSKDV Q QIDYKLES FQAVYNKLTG KQIVFEIPSE TH UniProtKB: Small ribosomal subunit protein eS7A |
+Macromolecule #9: rpS8_eS8
| Macromolecule | Name: rpS8_eS8 / type: protein_or_peptide / ID: 9 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 22.537803 KDa |
| Sequence | String: MGISRDSRHK RSATGAKRAQ FRKKRKFELG RQPANTKIGA KRIHSVRTRG GNKKYRALRI ETGNFSWASE GISKKTRIAG VVYHPSNNE LVRTNTLTKA AIVQIDATPF RQWFEAHYGQ TLGKKKNVKE EETVAKSKNA ERKWAARAAS AKIESSVESQ F SAGRLYAC ...String: MGISRDSRHK RSATGAKRAQ FRKKRKFELG RQPANTKIGA KRIHSVRTRG GNKKYRALRI ETGNFSWASE GISKKTRIAG VVYHPSNNE LVRTNTLTKA AIVQIDATPF RQWFEAHYGQ TLGKKKNVKE EETVAKSKNA ERKWAARAAS AKIESSVESQ F SAGRLYAC ISSRPGQSGR CDGYILEGEE LAFYLRRLTA KK UniProtKB: Small ribosomal subunit protein eS8A |
+Macromolecule #10: rpS9_uS4
| Macromolecule | Name: rpS9_uS4 / type: protein_or_peptide / ID: 10 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 22.487893 KDa |
| Sequence | String: MPRAPRTYSK TYSTPKRPYE SSRLDAELKL AGEFGLKNKK EIYRISFQLS KIRRAARDLL TRDEKDPKRL FEGNALIRRL VRVGVLSED KKKLDYVLAL KVEDFLERRL QTQVYKLGLA KSVHHARVLI TQRHIAVGKQ IVNIPSFMVR LDSEKHIDFA P TSPFGGAR ...String: MPRAPRTYSK TYSTPKRPYE SSRLDAELKL AGEFGLKNKK EIYRISFQLS KIRRAARDLL TRDEKDPKRL FEGNALIRRL VRVGVLSED KKKLDYVLAL KVEDFLERRL QTQVYKLGLA KSVHHARVLI TQRHIAVGKQ IVNIPSFMVR LDSEKHIDFA P TSPFGGAR PGRVARRNAA RKAEASGEAA DEADEADEE UniProtKB: Small ribosomal subunit protein uS4A |
+Macromolecule #11: rpS16_uS9
| Macromolecule | Name: rpS16_uS9 / type: protein_or_peptide / ID: 11 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 15.87749 KDa |
| Sequence | String: MSAVPSVQTF GKKKSATAVA HVKAGKGLIK VNGSPITLVE PEILRFKVYE PLLLVGLDKF SNIDIRVRVT GGGHVSQVYA IRQAIAKGL VAYHQKYVDE QSKNELKKAF TSYDRTLLIA DSRRPEPKKF GGKGARSRFQ KSYR UniProtKB: Small ribosomal subunit protein uS9A |
+Macromolecule #12: rpS11_uS17
| Macromolecule | Name: rpS11_uS17 / type: protein_or_peptide / ID: 12 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 17.785934 KDa |
| Sequence | String: MSTELTVQSE RAFQKQPHIF NNPKVKTSKR TKRWYKNAGL GFKTPKTAIE GSYIDKKCPF TGLVSIRGKI LTGTVVSTKM HRTIVIRRA YLHYIPKYNR YEKRHKNVPV HVSPAFRVQV GDIVTVGQCR PISKTVRFNV VKVSAAAGKA NKQFAKF UniProtKB: Small ribosomal subunit protein uS17A |
+Macromolecule #13: rpS22_uS8
| Macromolecule | Name: rpS22_uS8 / type: protein_or_peptide / ID: 13 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 14.650062 KDa |
| Sequence | String: MTRSSVLADA LNAINNAEKT GKRQVLIRPS SKVIIKFLQV MQKHGYIGEF EYIDDHRSGK IVVQLNGRLN KCGVISPRFN VKIGDIEKW TANLLPARQF GYVILTTSAG IMDHEEARRK HVSGKILGFV Y UniProtKB: Small ribosomal subunit protein uS8A |
+Macromolecule #14: rpS24_eS24
| Macromolecule | Name: rpS24_eS24 / type: protein_or_peptide / ID: 14 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 15.362848 KDa |
| Sequence | String: MSDAVTIRTR KVISNPLLAR KQFVVDVLHP NRANVSKDEL REKLAEVYKA EKDAVSVFGF RTQFGGGKSV GFGLVYNSVA EAKKFEPTY RLVRYGLAEK VEKASRQQRK QKKNRDKKIF GTGKRLAKKV ARRNAD UniProtKB: Small ribosomal subunit protein eS24A |
+Macromolecule #15: rpS28_eS28
| Macromolecule | Name: rpS28_eS28 / type: protein_or_peptide / ID: 15 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 7.605847 KDa |
| Sequence | String: MDNKTPVTLA KVIKVLGRTG SRGGVTQVRV EFLEDTSRTI VRNVKGPVRE NDILVLMESE REARRLR UniProtKB: Small ribosomal subunit protein eS28A |
+Macromolecule #16: Utp17
| Macromolecule | Name: Utp17 / type: protein_or_peptide / ID: 16 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 100.960445 KDa |
| Sequence | String: MTQSLGIEQY KLSVVSGGKP ALNNLSSVTG NKNIARLSQD QRNYIIPFNN QIKVYSVETR QCVKTLKFAN NSLLSGIFLQ EEENNESIV KILLGDITVP QQEDAHLITV FTNNGHVIVL NYKGKLVESP KHFKISLADE KLANVFHSEG NYRILTTFKD P SQKAHNSL ...String: MTQSLGIEQY KLSVVSGGKP ALNNLSSVTG NKNIARLSQD QRNYIIPFNN QIKVYSVETR QCVKTLKFAN NSLLSGIFLQ EEENNESIV KILLGDITVP QQEDAHLITV FTNNGHVIVL NYKGKLVESP KHFKISLADE KLANVFHSEG NYRILTTFKD P SQKAHNSL QSYRLYALTF DDAKKQFEVA HQAEWHNVIL SNISSNGKLL AHMCKDVSTK DHEHKSISVV SLFDDSVNLS FP LGSILSS QTQSLSYNTR YVSSMAIDNM GQQLAVGFAS GVISIVSLAD LQIRLLKWHI DSVLSLSFSH DGSYLLSGGW EKV MSLWQL ETNSQQFLPR LNGIIIDCQV LGPQGNYYSL ILQMTENNSN SDYQFLLLNA SDLTSKLSIN GPLPVFNSTI KHIQ QPISA MNTKNSNSIT SLNHSKKKQS RKLIKSRRQD FTTNVEINPI NKNLYFPHIS AVQIFDFYKN EQVNYQYLTS GVNNS MGKV RFELNLQDPI ITDLKFTKDG QWMITYEIEY PPNDLLSSKD LTHILKFWTK NDNETNWNLK TKVINPHGIS VPITKI LPS PRSVNNSQGC LTADNNGGLK FWSFDSHESN WCLKKISLPN FNHFSNSVSL AWSQDGSLIF HGFDDKLQIL DFDTFKK FE SLENTKTVSE FTLDSEIQTV KLINDTNLIV ATRTTLNAIN LLRGQVINSF DLYPFVNGVY KNGHMDRLIT CDERTGNI A LVINQQLTDL DGVPTINYKS RIIIFDSDLS TKLGNFTHHE YISWIGWNYD TDFIFLDIES TLGVVGTTVN TQLSDEVNN EGILDGLVSN TITTSAS(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)RGKKSDTR DKNTNDNDED EEDIALEFIN GEKKDKLVNM NSFTSMFDNI QNVQMDTFFD RVMKVLT UniProtKB: NET1-associated nuclear protein 1, NET1-associated nuclear protein 1 |
+Macromolecule #17: Utp8
| Macromolecule | Name: Utp8 / type: protein_or_peptide / ID: 17 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 65.572977 KDa |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) ...String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)RLFKQAIV TCPNLPLNEL LEELFSIRNR ELLLDISFRI LQDFTRDSIK QEMKKLSKLD VQ NFIEFIT SGGEDSSPEC FNPSQSTQLF QLLSLVLDSI GLFSLEGALL ENLTLYIDKQ VEIAERNTEL WNLIDTKGFQ HGF ASSTFD NGTSQKRALP TYTMEYLDI UniProtKB: U3 small nucleolar RNA-associated protein 8 |
+Macromolecule #18: Utp15
| Macromolecule | Name: Utp15 / type: protein_or_peptide / ID: 18 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 57.765289 KDa |
| Sequence | String: MSTARPRIIT SKAPLLPQQT TPEQRYWRQY TSAQLVKEHN SVTHISFNPQ HPHDFAVTSS TRVQIFSSRT RQVIKTFSRF KDVVYSASF RSDGKLLCAG DATGLVSVYD SYNPRTILLS INASTHPTHV TKFHTQDNKI LATASDDRVT RLWDISNAYE P QLELTGAT ...String: MSTARPRIIT SKAPLLPQQT TPEQRYWRQY TSAQLVKEHN SVTHISFNPQ HPHDFAVTSS TRVQIFSSRT RQVIKTFSRF KDVVYSASF RSDGKLLCAG DATGLVSVYD SYNPRTILLS INASTHPTHV TKFHTQDNKI LATASDDRVT RLWDISNAYE P QLELTGAT DYVRTLSFIP AAPHLVATGS YDGLIRLYDT RSSGSTPIYS LNHDQPVENV IAVSPTQIVS CGGNNFKVWD LT SNKKLYE RGNFNKAVTC LDYVENFDSP MQSALIASSL DGHVKVFDPL DNFQVKFGWK FSGPVLSCAV SPSTAQGNRH LVA GLSSGL LAIRTKKKEK RSSDKENAPA SFNKNAKSNN FQRMMRGSEY QGDQEHIIHN DKVRSQRRMR AFERNINQFK WSEA LDNAF VPGMAKELTL TVLQELRKRG KVRVALYGRD ESTLEPLLNW CLKGIEDVRS ASIVADWVAV VLELYGNTLE SSPVL QELM IDLKTKVRHE IHKSKEAQRI EGMLQLLTS UniProtKB: U3 small nucleolar RNA-associated protein 15 |
+Macromolecule #19: Utp9
| Macromolecule | Name: Utp9 / type: protein_or_peptide / ID: 19 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 64.378512 KDa |
| Sequence | String: MGSSLDLVAS FSHDSTRFAF QASVAQKNNV DIYPLNETKD YVVNSSLVSH IDYETNDMKV SDVIFFGWCS DLIDTQSSNI KRKLDEDEG TGESSEQRCE NFFVNGFPDG RIVVYSSNGK DIVNIIKNKK EILGADTDES DIWILDSDKV VKKLQYNNSK P LKTFTLVD ...String: MGSSLDLVAS FSHDSTRFAF QASVAQKNNV DIYPLNETKD YVVNSSLVSH IDYETNDMKV SDVIFFGWCS DLIDTQSSNI KRKLDEDEG TGESSEQRCE NFFVNGFPDG RIVVYSSNGK DIVNIIKNKK EILGADTDES DIWILDSDKV VKKLQYNNSK P LKTFTLVD GKDDEIVHFQ ILHQNGTLLV CIITKQMVYI VDPSKRRPST KYSFEISDAV ACEFSSDGKY LLIANNEELI AY DLKEDSK LIQSWPVQVK TLKTLDDLIM ALTTDGKINN YKIGEADKVC SIVVNEDLEI IDFTPINSKQ QVLISWLNVN EPN FESISL KEIETQGYIT INKNEKNNAD EADQKKLEEK EEEAQPEVQH EKKETETKIN KKVSKSDQVE IANILS(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)DHLSKI FLTISKSITQ NPWNEENLLP LWLKWLLTLK SGELNSIKDK HTKKNCKHLK SALRSSEEIL PVLL GIQGR LEMLRRQAKL REDLAQLSMQ EGEDDEIEVI EHSNVISNPL QDQASPVEKL EPDSIVYANG ESDEFVDASE YKD UniProtKB: U3 small nucleolar RNA-associated protein 9, U3 small nucleolar RNA-associated protein 9 |
+Macromolecule #20: Utp5
| Macromolecule | Name: Utp5 / type: protein_or_peptide / ID: 20 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 72.079445 KDa |
| Sequence | String: MDSPVLQSAY DPSGQYLCYV TVALDKQRVG VQPTQRATSS GVDTVWNENF LYLEDSKLKV TCLKWVNLAS SDTVAIILGM NNGEIWLYS VLANEVTYKF TTGNSYEIKD IDLMGNQLWC IDSSDAFYQF DLLQFKLLQH FRINNCVQLN KLTIVPAGDS V AQLLVASH ...String: MDSPVLQSAY DPSGQYLCYV TVALDKQRVG VQPTQRATSS GVDTVWNENF LYLEDSKLKV TCLKWVNLAS SDTVAIILGM NNGEIWLYS VLANEVTYKF TTGNSYEIKD IDLMGNQLWC IDSSDAFYQF DLLQFKLLQH FRINNCVQLN KLTIVPAGDS V AQLLVASH SISLIDIEEK KVVMTFPGHV SPVSTLQVIT NEFFISGAEG DRFLNVYDIH SGMTKCVLVA ESDIKELSHS GQ ADSIAVT TEDGSLEIFV DPLVSSSTKK RGNKSKKSSK KIQIVSKDGR KVPIYNAFIN KDLLNVSWLQ NATMPYFKNL QWR EIPNEY TVEISLNWNN KNKSADRDLH GKDLASATNY VEGNARVTSG DNFKHVDDAI KSWERELTSL EQEQAKPPQA NELL TETFG DKLESSTVAR ISGKKTNLKG SNLKTATTTG TVTVILSQAL QSNDHSLLET VLNNRDERVI RDTIFRLKPA LAVIL LERL AERIARQTHR QGPLNVWVKW CLIIHGGYLV SIPNLMSTLS SLHSTLKRRS DLLPRLLALD ARLDCTINKF KTLNYE AGD IHSSEPVVEE DEDDVEYNEE LDDAGLIEDG EESYGSEEEE EGDSDNEEEQ KHTSSKQDGR LETEQSDGEE EAGYSDV EM E UniProtKB: U3 small nucleolar RNA-associated protein 5 |
+Macromolecule #21: Utp10
| Macromolecule | Name: Utp10 / type: protein_or_peptide / ID: 21 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 200.020719 KDa |
| Sequence | String: MSSLSDQLAQ VASNNATVAL DRKRRQKLHS ASLIYNSKTA ATQDYDFIFE NASKALEELS QIEPKFAIFS RTLFSESSIS LDRNVQTKE EIKDLDNAIN AYLLLASSKW YLAPTLHATE WLVRRFQIHV KNTEMLLLST LNYYQTPVFK RILSIIKLPP L FNCLSNFV ...String: MSSLSDQLAQ VASNNATVAL DRKRRQKLHS ASLIYNSKTA ATQDYDFIFE NASKALEELS QIEPKFAIFS RTLFSESSIS LDRNVQTKE EIKDLDNAIN AYLLLASSKW YLAPTLHATE WLVRRFQIHV KNTEMLLLST LNYYQTPVFK RILSIIKLPP L FNCLSNFV RSEKPPTALT MIKLFNDMDF LKLYTSYLDQ CIKHNATYTN QLLFTTCCFI NVVAFNSNND EKLNQLVPIL LE ISAKLLA SKSKDCQIAA HTILVVFATA LPLKKTIILA AMETILSNLD AKEAKHSALL TICKLFQTLK GQGNVDQLPS KIF KLFDSK FDTVSILTFL DKEDKPVCDK FITSYTRSIA RYDRSKLNII LSLLKKIRLE RYEVRLIITD LIYLSEILED KSQL VELFE YFISINEDLV LKCLKSLGLT G(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)LTT SLFTNADVNT DIVKQLSDPV E TTKKDTAS FQTFLDKHSE LINTTNVSML TETGERYKKV LSLFTEAIGK GYKASSFLTS FFTTLESRIT FLLRVTISPA AP TALKLIS LNNIAKYINS IEKEVNIFTL VPCLICALRD ASIKVRTGVK KILSLIAKRP STKHYFLSDK LYGENVTIPM LNP KDSEAW LSGFLNEYVT ENYDISRILT PKRNEKVFLM FWANQALLIP SPYAKTVLLD NLNKSPTYAS SYSSLFEEFI SHYL ENRSS WEKSCIANKT NFEHFERSLV NLVSPKEKQS FMIDFVLSAL NSDYEQLANI AAERLISIFA SLNNAQKLKI VQNIV DSSS NVESSYDTVG VLQSLPLDSD IFVSILNQNS ISNEMDQTDF SKRRRRRSST SKNAFLKEEV SQLAELHLRK LTIILE ALD KVRNVGSEKL LFTLLSLLSD LETLDQDGGL PVLYAQETLI SCTLNTITYL KEHGCTELTN VRADILVSAI RNSASPQ VQ NKLLLVIGSL ATLSSEVILH SVMPIFTFMG AHSIRQDDEF TTKVVERTIL TVVPALIKNS KGNEKEEMEF LLLSFTTA L QHVPRHRRVK LFSTLIKTLD PVKALGSFLF LIAQQYSSAL VNFKIGEARI LIEFIKALLV DLHVNEELSG LNDLLDIIK LLTSSKSSSE KKKSLESRVL FSNGVLNFSE SEFLTFMNNT FEFINKITEE TDQDYYDVRR NLRLKVYSVL LDETSDKKLI RNIREEFGT LLEGVLFFIN SVELTFSCIT SQENEEASDS ETSLSDHTTE IKEILFKVLG NVLQILPVDE FVNAVLPLLS T STNEDIRY HLTLVIGSKF ELEGSEAIPI VNNVMKVLLD RMPLESKSVV ISQVILNTMT ALVSKYGKKL EGSILTQALT LA TEKVSSD MTEVKISSLA LITNCVQVLG VKSIAFYPKI VPPSIKLFDA SLADSSNPLK EQLQVAILLL FAGLIKRIPS FLM SNILDV LHVIYFSREV DSSIRLSVIS LIIENIDLKE VLKVLFRIWS TEIATSNDTV AVSLFLSTLE STVENIDKKS ATSQ SPIFF KLLLSLFEFR SISSFDNNTI SRIEASVHEI SNSYVLKMND KVFRPLFVIL VRWAFDGEGV TNAGITETER LLAFF KFFN KLQENLRGII TSYFTYLLEP VDMLLKRFIS KDMENVNLRR LVINSLTSSL KFDRDEYWKS TSRFELISVS LVNQLS NIE NSIGKYLVKA IGALASNNSG VDEHNQILNK LIVEHMKASC SSNEKLWAIR AMKLIYSKIG ESWLVLLPQL VPVIAEL LE DDDEEIEREV RTGLVKVVEN VLGEPFDRYL D UniProtKB: U3 small nucleolar RNA-associated protein 10, U3 small nucleolar RNA-associated protein 10 |
+Macromolecule #22: Utp4
| Macromolecule | Name: Utp4 / type: protein_or_peptide / ID: 22 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 87.3165 KDa |
| Sequence | String: MSSSLLSVLK EKSRSLKIRN KPVKMTSQER MIVHRCRFVD FTPATITSLA FSHKSNINKL TPSDLRLAIG RSNGNIEIWN PRNNWFQEM VIEGGKDRSI EGLCWSNVNG ESLRLFSIGG STVVTEWDLA TGLPLRNYDC NSGVIWSISI NDSQDKLSVG C DNGTVVLI ...String: MSSSLLSVLK EKSRSLKIRN KPVKMTSQER MIVHRCRFVD FTPATITSLA FSHKSNINKL TPSDLRLAIG RSNGNIEIWN PRNNWFQEM VIEGGKDRSI EGLCWSNVNG ESLRLFSIGG STVVTEWDLA TGLPLRNYDC NSGVIWSISI NDSQDKLSVG C DNGTVVLI DISGGPGVLE HDTILMRQEA RVLTLAWKKD DFVIGGCSDG RIRIWSAQKN DENMGRLLHT MKVDKAKKES TL VWSVIYL PRTDQIASGD STGSIKFWDF QFATLNQSFK AHDADVLCLT TDTDNNYVFS AGVDRKIFQF SQNTNKSQKN NRW VNSSNR LLHGNDIRAI CAYQSKGADF LVSGGVEKTL VINSLTSFSN GNYRKMPTVE PYSKNVLVNK EQRLVVSWSE STVK IWTMG TDSSTEQNYK LVCKLTLKDD QNISTCSLSP DGQVLVVGRP STTKVFHLQP VGNKLKVTKL DNDLLLRTST KLVKF IDNS KIVICSCEDD VFIVDLESEE DEKPQEVELL EVTSTKSSIK VPYINRINHL EVDQNIAVIS RGCGVVDILD LKARIS KPL ARLNNFITAV HINTSRKSVV VITADNKIYE FNMNLNSEAE NEDSESVLTQ WSKNNTDNLP KEWKTLKENC VGIFSDI EN SSRLWFWGAT WISRIDFDVD FPIN(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) TDESNFMNDE EDDEDDDIDM EISENLNVLL NQGNKIKSTD VQRNEESSGH FFFTD KYKP LLFVDLISSN ELAIIERNPL TFHSKQKAFI QPKLVF UniProtKB: U3 small nucleolar RNA-associated protein 4, U3 small nucleolar RNA-associated protein 4 |
+Macromolecule #23: Utp1
| Macromolecule | Name: Utp1 / type: protein_or_peptide / ID: 23 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 104.097039 KDa |
| Sequence | String: MKSDFKFSNL LGTVYRQGNI TFSDDGKQLL SPVGNRVSVF DLINNKSFTF EYEHRKNIAA IDLNKQGTLL ISIDEDGRAI LVNFKARNV LHHFNFKEKC SAVKFSPDGR LFALASGRFL QIWKTPDVNK DRQFAPFVRH RVHAGHFQDI TSLTWSQDSR F ILTTSKDL ...String: MKSDFKFSNL LGTVYRQGNI TFSDDGKQLL SPVGNRVSVF DLINNKSFTF EYEHRKNIAA IDLNKQGTLL ISIDEDGRAI LVNFKARNV LHHFNFKEKC SAVKFSPDGR LFALASGRFL QIWKTPDVNK DRQFAPFVRH RVHAGHFQDI TSLTWSQDSR F ILTTSKDL SAKIWSVDSE EKNLAATTFN GHRDYVMGAF FSHDQEKIYT VSKDGAVFVW EFTKRPSDDD DNESEDDDKQ EE VDISKYS WRITKKHFFY ANQAKVKCVT FHPATRLLAV GFTSGEFRLY DLPDFTLIQQ LSMGQNPVNT VSVNQTGEWL AFG SSKLGQ LLVYEWQSES YILKQQGHFD STNSLAYSPD GSRVVTASED GKIKVWDITS GFCLATFEEH TSSVTAVQFA KRGQ VMFSS SLDGTVRAWD LIRYRNFRTF TGTERIQFNC LAVDPSGEVV CAGSLDNFDI HVWSVQTGQL LDALSGHEGP VSCLS FSQE NSVLASASWD KTIRIWSIFG RSQQVEPIEV YSDVLALSMR PDGKEVAVST LKGQISIFNI EDAKQVGNID CRKDII SGR FNQDRFTAKN SERSKFFTTI HYSFDGMAIV AGGNNNSICL YDVPNEVLLK RFIVSRNMAL NGTLEFLNSK KMTEAGS LD LIDDAGENSD LEDRIDNSLP GSQRGGDLST RKMRPEVRVT SVQFSPTANA FAAASTEGLL IYSTNDTILF DPFDLDVD V TPHSTVEALR EKQFLNALVM AFRLNEEYLI NKVYEAIPIK EIPLVASNIP AIYLPRILKF IGDFAIESQH IEFNLIWIK ALLSASGGYI NEHKYLFSTA MRSIQRFIVR VAKEVVNTTT DNKYTYRFLV STDGSMEDGA ADDDEVLLKD DADEDNEENE ENDVVMESD DEEGWIGFNG KDNKLPLSNE NDSSDEEENE KELP UniProtKB: Periodic tryptophan protein 2 |
+Macromolecule #24: Utp6
| Macromolecule | Name: Utp6 / type: protein_or_peptide / ID: 24 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 48.437402 KDa |
| Sequence | String: MSKTRYYLEQ CIPEMDDLVE KGLFTKNEVS LIMKKRTDFE HRLNSRGSSI NDYIKYINYE SNVNKLRAKR CKRILQVKKT NSLSDWSIQ QRIGFIYQRG TNKFPQDLKF WAMYLNYMKA RGNQTSYKKI HNIYNQLLKL HPTNVDIWIS CAKYEYEVHA N FKSCRNIF ...String: MSKTRYYLEQ CIPEMDDLVE KGLFTKNEVS LIMKKRTDFE HRLNSRGSSI NDYIKYINYE SNVNKLRAKR CKRILQVKKT NSLSDWSIQ QRIGFIYQRG TNKFPQDLKF WAMYLNYMKA RGNQTSYKKI HNIYNQLLKL HPTNVDIWIS CAKYEYEVHA N FKSCRNIF QNGLRFNPDV PKLWYEYVKF ELNFITKLIN RRKVMGLINE REQELDMQNE QKNNQAPDEE KSHLQVPSTG DS MKDKLNE LPEADISVLG NAETNPALRG DIALTIFDVC MKTLGKHYIN KHKGYYAISD SKMNIELNKE TLNYLFSESL RYI KLFDEF (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK) UniProtKB: U3 small nucleolar RNA-associated protein 6 |
+Macromolecule #25: Utp12
| Macromolecule | Name: Utp12 / type: protein_or_peptide / ID: 25 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 105.037648 KDa |
| Sequence | String: MVKSYQRFEQ AAAFGVIASN ANCVWIPASS GNSNGSGPGQ LITSALEDVN IWDIKTGDLV SKLSDGLPPG ASDARGAKPA ECTYLEAHK DTDLLAVGYA DGVIKVWDLM SKTVLLNFNG HKAAITLLQF DGTGTRLISG SKDSNIIVWD LVGEVGLYKL R SHKDSITG ...String: MVKSYQRFEQ AAAFGVIASN ANCVWIPASS GNSNGSGPGQ LITSALEDVN IWDIKTGDLV SKLSDGLPPG ASDARGAKPA ECTYLEAHK DTDLLAVGYA DGVIKVWDLM SKTVLLNFNG HKAAITLLQF DGTGTRLISG SKDSNIIVWD LVGEVGLYKL R SHKDSITG FWCQGEDWLI STSKDGMIKL WDLKTHQCIE THIAHTGECW GLAVKDDLLI TTGTDSQVKI WKLDIENDKM GG KLTEMGI FEKQSKQRGL KIEFITNSSD KTSFFYIQNA DKTIETFRIR (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)YSSFI LHPFQTIRSL YKIKSASWTT VSSSKLELV LTTSSNTIEY YSIPYEKRDP TSPAPLKTHT IELQGQRTDV RSIDISDDNK LLATASNGSL KIWNIKTHKC I RTFECGYA LTCKFLPGGL LVILGTRNGE LQLFDLASSS LLDTIEDAHD AAIWSLDLTS DGKRLVTGSA DKTVKFWDFK VE NSLVPGT KNKFLPVLKL HHDTTLELTD DILCVRVSPD DRYLAISLLD NTVKVFFLDS MKFYLSLYGH KLPVLSIDIS FDS KMIITS SADKNIKIWG LDFGDCHKSL FAHQDSIMNV KFLPQSHNFF SCSKDAVVKY WDGEKFECIQ KLYAHQSEVW ALAV ATDGG FVVSSSHDHS IRIWEETEDQ VFLEEEKEKE LEEQYEDTLL TSLEEGNGDD AFKADASGEG VEDEASGVHK QTLES LKAG ERLMEALDLG IAEIEGLEAY NRDMKL(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)KP QGNAVLIAVN KTPEQYIMDT LLRIRMSQLE DALMVMPFSY VLKFLKFIDT VMQNKTLLHS HLPLICKNLF FII KFNHKE LVSQKNEELK LQINRVKTEL RSALKSTEDD LGFNVQGLKF VKQQWNLRHN YEFVDEYDQQ EKESNSARKR VFGT VI UniProtKB: U3 small nucleolar RNA-associated protein 12, U3 small nucleolar RNA-associated protein 12, U3 small nucleolar RNA-associated protein 12 |
+Macromolecule #26: Utp13
| Macromolecule | Name: Utp13 / type: protein_or_peptide / ID: 26 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 75.035867 KDa |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) ...String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)EQ EIEEEQEKAK LQVEQEQSLQ NYMSKGDWTN AFLLAMTL D HPMRLFNVLK RALGESRSRQ DTEEGKIEVI FNEELDQAIS ILNDEQLILL MKRCRDWNTN AKTHTIAQRT IRCILMHHN IAKLSEIPGM VKIVDAIIPY TQRHFTRVDN LVEQSYILDY ALVEMDKLF UniProtKB: U3 small nucleolar RNA-associated protein 13 |
+Macromolecule #27: Utp18
| Macromolecule | Name: Utp18 / type: protein_or_peptide / ID: 27 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 66.49425 KDa |
| Sequence | String: MTMATTAMNV SVPPPDEEEQ LLAKFVFGDT TDLQENLAKF NADFIFNEQE MDVEDQEDEG SESDNSEEDE AQNGELDHVN NDQLFFVDD GGNEDSQDKN EDTMDVDDED DSSSDDYSED SEEAAWIDSD DEKIKVPILV TNKTKKLRTS YNESKINGVH Y INRLRSQF ...String: MTMATTAMNV SVPPPDEEEQ LLAKFVFGDT TDLQENLAKF NADFIFNEQE MDVEDQEDEG SESDNSEEDE AQNGELDHVN NDQLFFVDD GGNEDSQDKN EDTMDVDDED DSSSDDYSED SEEAAWIDSD DEKIKVPILV TNKTKKLRTS YNESKINGVH Y INRLRSQF EKIYPRPKWV DDESDSELDD EEDDEEEGSN NVINGDINAL TKILSTTYNY KDTLSNSKLL PPKKLDIVRL KD ANASHPS HSAIQSLSFH PSKPLLLTGG YDKTLRIYHI DGKTNHLVTS LHLVGSPIQT CTFYTSLSNQ NQQNIFTAGR RRY MHSWDL SLENLTHSQT AKIEKFSRLY GHESTQRSFE NFKVAHLQNS QTNSVHGIVL LQGNNGWINI LHSTSGLWLM GCKI EGVIT DFCIDYQPIS RGKFRTILIA VNAYGEVWEF DLNKNGHVIR RWKDQGGVGI TKIQVGGGTT TTCPALQISK IKQNR WLAV GSESGFVNLY DRNNAMTSST PTPVAALDQL TTTISNLQFS PDGQILCMAS RAVKDALRLV HLPSCSVFSN WPTSGT PLG KVTSVAFSPS GGLLAVGNEQ GKVRLWKLNH Y UniProtKB: U3 small nucleolar RNA-associated protein 18 |
+Macromolecule #28: Utp21
| Macromolecule | Name: Utp21 / type: protein_or_peptide / ID: 28 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 104.927844 KDa |
| Sequence | String: MSIDLKKRKV EEDVRSRGKN SKIFSPFRII GNVSNGVPFA TGTLGSTFYI VTCVGKTFQI YDANTLHLLF VSEKETPSSI VALSAHFHY VYAAYENKVG IYKRGIEEHL LELETDANVE HLCIFGDYLC ASTDDNSIFI YKKSDPQDKY PSEFYTKLTV T EIQGGEIV ...String: MSIDLKKRKV EEDVRSRGKN SKIFSPFRII GNVSNGVPFA TGTLGSTFYI VTCVGKTFQI YDANTLHLLF VSEKETPSSI VALSAHFHY VYAAYENKVG IYKRGIEEHL LELETDANVE HLCIFGDYLC ASTDDNSIFI YKKSDPQDKY PSEFYTKLTV T EIQGGEIV SLQHLATYLN KLTVVTKSNV LLFNVRTGKL VFTSNEFPDQ ITTAEPAPVL DIIALGTVTG EVIMFNMRKG KR IRTIKIP QSRISSLSFR TDGSSHLSVG TSSGDLIFYD LDRRSRIHVL KNIHRESYGG VTQATFLNGQ PIIVTSGGDN SLK EYVFDP SLSQGSGDVV VQPPRYLRSR GGHSQPPSYI AFADSQSHFM LSASKDRSLW SFSLRKDAQS QEMSQRLHKK QDGG RVGGS TIKSKFPEIV ALAIENARIG EWENIITAHK DEKFARTWDM RNKRVGRWTF DTTDDGFVKS VAMSQCGNFG FIGSS NGSI TIYNMQSGIL RKKYKLHKRA VTGISLDGMN RKMVSCGLDG IVGFYDFNKS TLLGKLKLDA PITAMVYHRS SDLFAL ALD DLSIVVIDAV TQRVVRQLWG HSNRITAFDF SPEGRWIVSA SLDSTIRTWD LPTGGCIDGI IVDNVATNVK FSPNGDL LA TTHVTGNGIC IWTNRAQFKT VSTRTIDESE FARMALPSTS VRGNDSMLSG ALESNGGEDL NDIDFNTYTS LEQIDKEL L TLSIGPRSKM NTLLHLDVIR KRSKPKEAPK KSEKLPFFLQ LSGEKVGDEA SVREGIAHET PEEIHRRDQE AQKKLDAEE QMNKFKVTGR LGFESHFTKQ LREGSQSKDY SSLLATLINF SPAAVDLEIR SLNSFEPFDE IVWFIDALTQ GLKSNKNFEL YETFMSLLF KAHGDVIHAN NKNQDIASAL QNWEDVHKKE DRLDDLVKFC MGVAAFVTTA UniProtKB: U3 small nucleolar RNA-associated protein 21 |
+Macromolecule #29: Sof1
| Macromolecule | Name: Sof1 / type: protein_or_peptide / ID: 29 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 56.888918 KDa |
| Sequence | String: MKIKTIKRSA DDYVPVKSTQ ESQMPRNLNP ELHPFERARE YTKALNATKL ERMFAKPFVG QLGYGHRDGV YAIAKNYGSL NKLATGSAD GVIKYWNMST REEFVSFKAH YGLVTGLCVT QPRFHDKKPD LKSQNFMLSC SDDKTVKLWS INVDDYSNKN S SDNDSVTN ...String: MKIKTIKRSA DDYVPVKSTQ ESQMPRNLNP ELHPFERARE YTKALNATKL ERMFAKPFVG QLGYGHRDGV YAIAKNYGSL NKLATGSAD GVIKYWNMST REEFVSFKAH YGLVTGLCVT QPRFHDKKPD LKSQNFMLSC SDDKTVKLWS INVDDYSNKN S SDNDSVTN EEGLIRTFDG ESAFQGIDSH RENSTFATGG AKIHLWDVNR LKPVSDLSWG ADNITSLKFN QNETDILAST GS DNSIVLY DLRTNSPTQK IVQTMRTNAI CWNPMEAFNF VTANEDHNAY YYDMRNLSRS LNVFKDHVSA VMDVDFSPTG DEI VTGSYD KSIRIYKTNH GHSREIYHTK RMQHVFQVKY SMDSKYIISG SDDGNVRLWR SKAWERSNVK TTREKNKLEY DEKL KERFR HMPEIKRISR HRHVPQVIKK AQEIKNIELS SIKRREANER RTRKDMPYIS ERKKQIVGTV HKYEDSGRDR KRRKE DDKR DTQEK UniProtKB: Protein SOF1 |
+Macromolecule #30: Enp2
| Macromolecule | Name: Enp2 / type: protein_or_peptide / ID: 30 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 81.864172 KDa |
| Sequence | String: MVLKSTSAND VSVYQVSGTN VSRSLPDWIA KKRKRQLKND LEYQNRVELI QDFEFSEASN KIKVSRDGQY CMATGTYKPQ IHVYDFANL SLKFDRHTDA ENVDFTILSD DWTKSVHLQN DRSIQFQNKG GLHYTTRIPK FGRSLVYNKV NCDLYVGASG N ELYRLNLE ...String: MVLKSTSAND VSVYQVSGTN VSRSLPDWIA KKRKRQLKND LEYQNRVELI QDFEFSEASN KIKVSRDGQY CMATGTYKPQ IHVYDFANL SLKFDRHTDA ENVDFTILSD DWTKSVHLQN DRSIQFQNKG GLHYTTRIPK FGRSLVYNKV NCDLYVGASG N ELYRLNLE KGRFLNPFKL DTEGVNHVSI NEVNGLLAAG TETNVVEFWD PRSRSRVSKL YLENNIDNRP FQVTTTSFRN DG LTFACGT SNGYSYIYDL RTSEPSIIKD QGYGFDIKKI IWLDNVGTEN KIVTCDKRIA KIWDRLDGKA YASMEPSVDI NDI EHVPGT GMFFTANESI PMHTYYIPSL GPSPRWCSFL DSITEELEEK PSDTVYSNYR FITRDDVKKL NLTHLVGSRV LRAY MHGFF INTELYDKVS LIANPDAYKD EREREIRRRI EKERESRIRS SGAVQKPKIK VNKTLVDKLS QKRGDKVAGK VLTDD RFKE MFEDEEFQVD EDDYDFKQLN PVKSIKETEE GAAKRIRALT AAEESDEERI AMKDGRGHYD YEDEESDEEE SDDETN QKS NKEELSEKDL RKMEKQKALI ERRKKEKEQS ERFMNEMKAG TSTSTQRDES AHVTFGEQVG ELLEVENGKK SNESILR RN QRGEAELTFI PQRKSKKDGN YKSRRHDNSS DEEGIDENGN KKDNGRSKPR FENRRRASKN AFRGM UniProtKB: Ribosome biogenesis protein ENP2 |
+Macromolecule #31: Utp7
| Macromolecule | Name: Utp7 / type: protein_or_peptide / ID: 31 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 62.41857 KDa |
| Sequence | String: MGHKKNGHRR QIKERENQNK FERSTYTNNA KNNHTQTKDK KLRAGLKKID EQYKKAVSSA AATDYLLPES NGYLEPENEL EKTFKVQQS EIKSSVDVST ANKALDLSLK EFGPYHIKYA KNGTHLLITG RKGHVASMDW RKGQLRAELF LNETCHSATY L QNEQYFAV ...String: MGHKKNGHRR QIKERENQNK FERSTYTNNA KNNHTQTKDK KLRAGLKKID EQYKKAVSSA AATDYLLPES NGYLEPENEL EKTFKVQQS EIKSSVDVST ANKALDLSLK EFGPYHIKYA KNGTHLLITG RKGHVASMDW RKGQLRAELF LNETCHSATY L QNEQYFAV AQKKYTFIYD HEGTELHRLK QHIEARHLDF LPYHYLLVTA GETGWLKYHD VSTGQLVSEL RTKAGPTMAM AQ NPWNAVM HLGHSNGTVS LWSPSMPEPL VKLLSARGPV NSIAIDRSGY YMATTGADRS MKIWDIRNFK QLHSVESLPT PGT NVSISD TGLLALSRGP HVTLWKDALK LSGDSKPCFG SMGGNPHRNT PYMSHLFAGN KVENLGFVPF EDLLGVGHQT GITN LIVPG AGEANYDALE LNPFETKKQR QEQEVRTLLN KLPADTITLD PNSIGSVDKR SSTIRLNAKD LAQTTMDANN KAKTN SDIP DVKPDVKGKN SGLRSFLRKK TQNVIDERKL RVQKQLDKEK NIRKRNHQIK QGLISEDHKD VIEEALSRFG UniProtKB: U3 small nucleolar RNA-associated protein 7 |
+Macromolecule #32: Kre33
| Macromolecule | Name: Kre33 / type: protein_or_peptide / ID: 32 / Number of copies: 2 / Enantiomer: LEVO EC number: Transferases; Acyltransferases; Transferring groups other than aminoacyl groups |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 95.337062 KDa |
| Sequence | String: MAKKAIDSRI PSLIRNGVQT KQRSIFVIVG DRARNQLPNL HYLMMSADLK MNKSVLWAYK KKLLGFTSHR KKRENKIKKE IKRGTREVN EMDPFESFIS NQNIRYVYYK ESEKILGNTY GMCILQDFEA LTPNLLARTI ETVEGGGIVV ILLKS(UNK) (UNK)(UNK)(UNK) ...String: MAKKAIDSRI PSLIRNGVQT KQRSIFVIVG DRARNQLPNL HYLMMSADLK MNKSVLWAYK KKLLGFTSHR KKRENKIKKE IKRGTREVN EMDPFESFIS NQNIRYVYYK ESEKILGNTY GMCILQDFEA LTPNLLARTI ETVEGGGIVV ILLKS(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)VVARFNER FILSLGSNPN CLVVDDELNV L(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) UniProtKB: RNA cytidine acetyltransferase, RNA cytidine acetyltransferase |
+Macromolecule #33: Imp3
| Macromolecule | Name: Imp3 / type: protein_or_peptide / ID: 33 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 21.928529 KDa |
| Sequence | String: MVRKLKHHEQ KLLKKVDFLE WKQDQGHRDT QVMRTYHIQN REDYHKYNRI CGDIRRLANK LSLLPPTDPF RRKHEQLLLD KLYAMGVLT TKSKISDLEN KVTVSAICRR RLPVIMHRLK MAETIQDAVK FIEQGHVRVG PNLINDPAYL VTRNMEDYVT W VDNSKIKK TLLRYRNQID DFDFS UniProtKB: U3 small nucleolar ribonucleoprotein protein IMP3 |
+Macromolecule #34: Mpp10
| Macromolecule | Name: Mpp10 / type: protein_or_peptide / ID: 34 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 67.042492 KDa |
| Sequence | String: MSELFGVLKS NAGRIILKDP SATSKDVKAY IDSVINTCKN GSITKKAELD EITVDGLDAN QVWWQVKLVL DSIDGDLIQG IQELKDVVT PSHNLSDGST LNSSSGEESE LEEAESVFKE KQMLSADVSE IEEQSNDSLS ENDEEPSMDD EKTSAEAARE E FAEEKRIS ...String: MSELFGVLKS NAGRIILKDP SATSKDVKAY IDSVINTCKN GSITKKAELD EITVDGLDAN QVWWQVKLVL DSIDGDLIQG IQELKDVVT PSHNLSDGST LNSSSGEESE LEEAESVFKE KQMLSADVSE IEEQSNDSLS ENDEEPSMDD EKTSAEAARE E FAEEKRIS SGQDERHSSP DPYGINDKFF DLEKFNRDTL AAEDSNEASE GSEDEDIDYF QDMPSDDEEE EAIYYEDFFD KP TKEPVKK HSDVKDPKED EELDEEEHDS AMDKVKLDLF ADEEDEPNAE GVGEASDKNL SSFEKQQIEI RKQIEQLENE AVA EKKWSL KGEVKAKDRP EDALLTEELE FDRTAKPVPV ITSEVTESLE DMIRRRIQDS NFDDLQRRTL LDITRKSQRP QFEL SDVKS SKSLAEIYED DYTRAEDESA LSEELQKAHS EISELYANLV YKLDVLSSVH FVPKPASTSL EIRVETPTIS MEDAQ PLYM SNASSLAPQE IYNVGKAEKD GEIRLKNGVA MSKEELTRED KNRLRRALKR KRSKANLPNV NKRSKRNDVV DTLSKA KNI TVINQKGEKK DVSGKTKKSR SGPDSTNIKL UniProtKB: U3 small nucleolar RNA-associated protein MPP10 |
+Macromolecule #35: Sas10
| Macromolecule | Name: Sas10 / type: protein_or_peptide / ID: 35 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 69.705703 KDa |
| Sequence | String: MVRKGSNRTK TSEVGDEINP YGLNEVDDFA SKREKVLLGQ STFGDSNKDD DHSLLEDEDE EEVLAMDEDD ESIDEREDEE EEEEEELDG AAAYKKIFGR NLETDQLPEE DEENGMLDNE NAWGSTKGEY YGADDLDDDE AAKEIEKEAL RQQKKHLEEL N MNDYLDEE ...String: MVRKGSNRTK TSEVGDEINP YGLNEVDDFA SKREKVLLGQ STFGDSNKDD DHSLLEDEDE EEVLAMDEDD ESIDEREDEE EEEEEELDG AAAYKKIFGR NLETDQLPEE DEENGMLDNE NAWGSTKGEY YGADDLDDDE AAKEIEKEAL RQQKKHLEEL N MNDYLDEE EEEEWVKSAK EFDMGEFKNS TKQADTKTSI TDILNMDDEA RDNYLRTMFP EFAPLSKEFT ELAPKFDELK KS EENEFNK LKLIALGSYL GTISCYYSIL LHELHNNEDF TSMKGHPVME KILTTKEIWR QASELPSSFD VNEGDGSESE ETA NIEAFN EKKLNELQNS EDSDAEDGGK QKQEIDEEER ESDEEEEEED VDIDDFEEYV AQSRLHSKPK TSSMPEADDF IESE IADVD AQDKKARRRT LRFYTSKIDQ QENKKTDRFK GDDDIP(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) DEARKRGMHD NNGADLDDKD YGSEDEAVSR SINTQGENDY YQQVQRGKQD KKISRKEAH KNAVIAAREG KLAELAENVS GDGKRAINYQ ILKNKGLTPK RNKDNRNSRV KKRKKYQKAQ KKLKSVRAVY S GGQSGVYE GEKTGIKKGL TRSVKFKN UniProtKB: Something about silencing protein 10, Something about silencing protein 10 |
+Macromolecule #36: Lcp5
| Macromolecule | Name: Lcp5 / type: protein_or_peptide / ID: 36 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 40.864391 KDa |
| Sequence | String: MSELNALLKD INGSLTATSE SLERLSGIYS NSATDEIPES NQLHEHLFYD AKKPAEKVSL LSLKNGSMLG YINSLLMLIG NRLDDECKD PSAMDARERS IQHRVVLERG VKPLEKKLAY QLDKLTRAYV KMEKEYKDAE KRALEKSTLV NHSGNDDSED D ESSEDEIA ...String: MSELNALLKD INGSLTATSE SLERLSGIYS NSATDEIPES NQLHEHLFYD AKKPAEKVSL LSLKNGSMLG YINSLLMLIG NRLDDECKD PSAMDARERS IQHRVVLERG VKPLEKKLAY QLDKLTRAYV KMEKEYKDAE KRALEKSTLV NHSGNDDSED D ESSEDEIA YRPNTSGIIN TNKKSSAYRV EETAKQENGE ENDDNETGVY KPPKITAVLP PQQTHFEDRF DAREHKDRSN KS RMQAMEE YIRESSDQPD WSASIGADIV NHGRGGIKSL RDTEKERRVT SFEEDNFTRL NITNKAEKRK QKQRERNARM NVI GGEDFG IFSSKRKLED STSRRGAKKT RSAWDRAQRR L UniProtKB: U3 small nucleolar ribonucleoprotein protein LCP5 |
+Macromolecule #37: Bud21
| Macromolecule | Name: Bud21 / type: protein_or_peptide / ID: 37 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 23.835627 KDa |
| Sequence | String: MSNGHVKFDA DESQASASAV TDRQDDVLVI SKKDKEVHSS SDEESDDDDA PQEEGLHSGK SEVESQITQR EEAIRLEQSQ LRSKRRKQN ELYAKQKKSV N(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK) ...String: MSNGHVKFDA DESQASASAV TDRQDDVLVI SKKDKEVHSS SDEESDDDDA PQEEGLHSGK SEVESQITQR EEAIRLEQSQ LRSKRRKQN ELYAKQKKSV N(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)QKDEGST QYSSSRHVTF DKLDESDENE EALAKAIK T KKRKTLKNLR KDSVKRGKFR VQLLSTTQDS KTLPPKKESS IIRSKDRWLN RKALNKG UniProtKB: U3 small nucleolar RNA-associated protein 16, U3 small nucleolar RNA-associated protein 16 |
+Macromolecule #38: Faf1
| Macromolecule | Name: Faf1 / type: protein_or_peptide / ID: 38 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 38.173699 KDa |
| Sequence | String: MTLDDDDYIK QMELQRKAFE SQFGSLESMG FEDKTKNIRT EVDTRDSSGD EIDNSDHGSD FKDGTIESSN SSDEDSGNET AEENNQDSK PK(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) ...String: MTLDDDDYIK QMELQRKAFE SQFGSLESMG FEDKTKNIRT EVDTRDSSGD EIDNSDHGSD FKDGTIESSN SSDEDSGNET AEENNQDSK PK(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) SGKTLTQINK KLESTEAKE EKEDETLEAE NLQNDLELQQ FLRESHLLSA FNNGGSGSTN SGVSLTLQSM GGGNDDGIVY QDDQVIGKAR S RTLEMRLN RLSRVNGHQD KINKLEKVPM HIRRGMIDKH VKRIKKYEQE AAEGGIVLSK VKKGQFRKIE STYKKDIERR IG GSIKARD KEKATKRERG LKISSVGRST RNGLIVSKRD IARISGGERS GKFNGKKKSR R UniProtKB: Protein FAF1, Protein FAF1 |
+Macromolecule #39: rpS13_uS15
| Macromolecule | Name: rpS13_uS15 / type: protein_or_peptide / ID: 39 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 17.059945 KDa |
| Sequence | String: MGRMHSAGKG ISSSAIPYSR NAPAWFKLSS ESVIEQIVKY ARKGLTPSQI GVLLRDAHGV TQARVITGNK IMRILKSNGL APEIPEDLY YLIKKAVSVR KHLERNRKDK DAKFRLILIE SRIHRLARYY RTVAVLPPNW KYESATASAL VN UniProtKB: Small ribosomal subunit protein uS15 |
+Macromolecule #40: rpS14_uS11
| Macromolecule | Name: rpS14_uS11 / type: protein_or_peptide / ID: 40 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 14.562655 KDa |
| Sequence | String: MSNVVQARDN SQVFGVARIY ASFNDTFVHV TDLSGKETIA RVTGGMKVKA DRDESSPYAA MLAAQDVAAK CKEVGITAVH VKIRATGGT RTKTPGPGGQ AALRALARSG LRIGRIEDVT PVPSDSTRKK GGRRGRRL UniProtKB: Small ribosomal subunit protein uS11A |
+Macromolecule #41: Utp22
| Macromolecule | Name: Utp22 / type: protein_or_peptide / ID: 41 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 140.660141 KDa |
| Sequence | String: MATSVKRKAS ETSDQNIVKV QKKHSTQDST TDNGSKENDH SSQAINERTV PEQENDESDT SPESNEVATN TAATRHNGKV TATESYDIH IARETAELFK SNIFKLQIDE LLEQVKLKQK HVLKVEKFLH KLYDILQEIP DWEEKSLAEV DSFFKNKIVS V PFVDPKPI ...String: MATSVKRKAS ETSDQNIVKV QKKHSTQDST TDNGSKENDH SSQAINERTV PEQENDESDT SPESNEVATN TAATRHNGKV TATESYDIH IARETAELFK SNIFKLQIDE LLEQVKLKQK HVLKVEKFLH KLYDILQEIP DWEEKSLAEV DSFFKNKIVS V PFVDPKPI PQNTNYKFNY KKPDISLIGS FALKAGIYQP NGSSIDTLLT MPKELFEKKD FLNFRCLHKR SVYLAYLTHH LL ILLKKDK LDSFLQLEYS YFDNDPLLPI LRISCSKPTG DSLSDYNFYK TRFSINLLIG FPYKVFEPKK LLPNRNCIRI AQE SKEQSL PATPLYNFSV LSSSTHENYL KYLYKTKKQT ESFVEATVLG RLWLQQRGFS SNMSHSGSLG GFGTFEFTIL MAAL LNGGG INSNKILLHG FSSYQLFKGV IKYLATMDLC HDGHLQFHSN PENSSSSPAS KYIDEGFQTP TLFDKSTKVN ILTKM TVSS YQILKEYAGE TLRMLNNVVQ DQFSNIFLTN ISRFDNLKYD LCYDVQLPLG KYNNLETSLA ATFGSMERVK FITLEN FLA HKITNVARYA LGDRIKYIQI EMVGQKSDFP ITKRKVYSNT GGNHFNFDFV RVKLIVNPSE CDKLVTKGPA HSETMST EA AVFKNFWGIK SSLRRFKDGS ITHCCVWSTS SSEPIISSIV NFALQKHVSK KAQISNETIK KFHNFLPLPN LPSSAKTS V LNLSSFFNLK KSFDDLYKII FQMKLPLSVK SILPVGSAFR YTSLCQPVPF AYSDPDFFQD VILEFETSPK WPDEITSLE KAKTAFLLKI QEELSANSST YRSFFSRDES IPYNLEIVTL NILTPEGYGF KFRVLTERDE ILYLRAIANA RNELKPELEA TFLKFTAKY LASVRHTRTL ENISHSYQFY SPVVRLFKRW LDTHLLLGHI TDELAELIAI KPFVDPAPYF IPGSLENGFL K VLKFISQW NWKDDPLILD LVKPEDDIRD TFETSIGAGS ELDSKTMKKL SERLTLAQYK GIQMNFTNLR NSDPNGTHLQ FF VASKNDP SGILYSSGIP LPIATRLTAL AKVAVNLLQT HGLNQQTINL LFTPGLKDYD FVVDLRTPIG LKSSCGILSA TEF KNITND QAPSNFPENL NDLSEKMDPT YQLVKYLNLK YKNSLILSSR KYIGVNGGEK GDKNVITGLI KPLFKGAHKF RVNL DCNVK PVDDENVILN KEAIFHEIAA FGNDMVINFE TD UniProtKB: U3 small nucleolar RNA-associated protein 22 |
+Macromolecule #42: Rrp7
| Macromolecule | Name: Rrp7 / type: protein_or_peptide / ID: 42 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 34.526441 KDa |
| Sequence | String: MGIEDISAMK NGFIVVPFKL PDHKALPKSQ EASLHFMFAK RHQSSNSNES DCLFLVNLPL LSNIEHMKKF VGQLCGKYDT VSHVEELLY NDEFGLHEVD LSALTSDLMS STDVNEKRYT PRNTALLKFV DAASINNCWN ALKKYSNLHA KHPNELFEWT Y TTPSFTTF ...String: MGIEDISAMK NGFIVVPFKL PDHKALPKSQ EASLHFMFAK RHQSSNSNES DCLFLVNLPL LSNIEHMKKF VGQLCGKYDT VSHVEELLY NDEFGLHEVD LSALTSDLMS STDVNEKRYT PRNTALLKFV DAASINNCWN ALKKYSNLHA KHPNELFEWT Y TTPSFTTF VNFYKPLDID YLKEDIHTHM AIFEQREAQA QEDVQSSIVD EDGFTLVVGK NTKSLNSIRK KILNKNPLSK HE NKAKPIS NIDKKAKKDF YRFQVRERKK QEINQLLSKF KEDQERIKVM KAKRKFNPYT UniProtKB: Ribosomal RNA-processing protein 7 |
+Macromolecule #43: Rrp5
| Macromolecule | Name: Rrp5 / type: protein_or_peptide / ID: 43 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 193.411422 KDa |
| Sequence | String: MVASTKRKRD EDFPLSREDS TKQPSTSSLV RNTEEVSFPR GGASALTPLE LKQVANEAAS DVLFGNESVK ASEPASRPLK KKKTTKKST SKDSEASSAN SDEARAGLIE HVNFKTLKNG SSLLGQISAI TKQDLCITFT DGISGYVNLT HISEEFTSIL E DLDEDMDS ...String: MVASTKRKRD EDFPLSREDS TKQPSTSSLV RNTEEVSFPR GGASALTPLE LKQVANEAAS DVLFGNESVK ASEPASRPLK KKKTTKKST SKDSEASSAN SDEARAGLIE HVNFKTLKNG SSLLGQISAI TKQDLCITFT DGISGYVNLT HISEEFTSIL E DLDEDMDS DTDAADEKKS KVEDAEYESS DDEDEKLDKS NELPNLRRYF HIGQWLRCSV IKNTSLEPST KKSKKKRIEL TI EPSSVNI YADEDLVKST SIQCAVKSIE DHGATLDVGL PGFTGFIAKK DFGNFEKLLP GAVFLGNITK KSDRSIVVNT DFS DKKNKI TQISSIDAII PGQIVDLLCE SITKNGIAGK VFGLVSGVVN VSHLRTFSEE DLKHKFVIGS SIRCRIIACL ENKS GDKVL ILSNLPHILK LEDALRSTEG LDAFPIGYTF ESCSIKGRDS EYLYLALDDD RLGKVHSSRV GEIENSENLS SRVLG YSPV DDIYQLSTDP KYLKLKYLRT NDIPIGELLP SCEITSVSSS GIELKIFNGQ FKASVPPLHI SDTRLVYPER KFKIGS KVK GRVISVNSRG NVHVTLKKSL VNIEDNELPL VSTYENAKNI KEKNEKTLAT IQVFKPNGCI ISFFGGLSGF LPNSEIS EV FVKRPEEHLR LGQTVIVKLL DVDADRRRII ATCKVSNEQA AQQKDTIENI VPGRTIITVH VIEKTKDSVI VEIPDVGL R GVIYVGHLSD SRIEQNRAQL KKLRIGTELT GLVIDKDTRT RVFNMSLKSS LIKDAKKETL PLTYDDVKDL NKDVPMHAY IKSISDKGLF VAFNGKFIGL VLPSYAVDSR DIDISKAFYI NQSVTVYLLR TDDKNQKFLL SLKAPKVKEE KKKVESNIED PVDSSIKSW DDLSIGSIVK AKIKSVKKNQ LNVILAANLH GRVDIAEVFD TYEEITDKKQ PLSNYKKDDV IKVKIIGNHD V KSHKFLPI THKISKASVL ELSMKPSELK SKEVHTKSLE EINIGQELTG FVNNSSGNHL WLTISPVLKA RISLLDLADN DS NFSENIE SVFPLGSALQ VKVASIDREH GFVNAIGKSH VDINMSTIKV GDELPGRVLK IAEKYVLLDL GNKVTGISFI TDA LNDFSL TLKEAFEDKI NNVIPTTVLS VDEQNKKIEL SLRPATAKTR SIKSHEDLKQ GEIVDGIVKN VNDKGIFVYL SRKV EAFVP VSKLSDSYLK EWKKFYKPMQ YVLGKVVTCD EDSRISLTLR ESEINGDLKV LKTYSDIKAG DVFEGTIKSV TDFGV FVKL DNTVNVTGLA HITEIADKKP EDLSALFGVG DRVKAIVLKT NPEKKQISLS LKASHFSKEA ELASTTTTTT TVDQLE KED EDEVMADAGF NDSDSESDIG DQNTEVADRK PETSSDGLSL SAGFDWTASI LDQAQEEEES DQDQEDFTEN KKHKHKR RK ENVVQDKTID INTRAPESVA DFERLLIGNP NSSVVWMNYM AFQLQLSEIE KARELAERAL KTINFREEAE KLNIWIAM L NLENTFGTEE TLEEVFSRAC QYMDSYTIHT KLLGIYEISE KFDKAAELFK ATAKKFGGEK VSIWVSWGDF LISHNEEQE ARTILGNALK ALPKRNHIEV VRKFAQLEFA KGDPERGRSL FEGLVADAPK RIDLWNVYVD QEVKAKDKKK VEDLFERIIT KKITRKQAK FFFNKWLQFE ESEGDEKTIE YVKAKATEYV ASHESQKADE UniProtKB: rRNA biogenesis protein RRP5 |
+Macromolecule #44: Krr1
| Macromolecule | Name: Krr1 / type: protein_or_peptide / ID: 44 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 37.226254 KDa |
| Sequence | String: MVSTHNRDKP WDTDDIDKWK IEEFKEEDNA SGQPFAEESS FMTLFPKYRE SYLKTIWNDV TRALDKHNIA CVLDLVEGSM TVKTTRKTY DPAIILKARD LIKLLARSVP FPQAVKILQD DMACDVIKIG NFVTNKERFV KRRQRLVGPN GNTLKALELL T KCYILVQG ...String: MVSTHNRDKP WDTDDIDKWK IEEFKEEDNA SGQPFAEESS FMTLFPKYRE SYLKTIWNDV TRALDKHNIA CVLDLVEGSM TVKTTRKTY DPAIILKARD LIKLLARSVP FPQAVKILQD DMACDVIKIG NFVTNKERFV KRRQRLVGPN GNTLKALELL T KCYILVQG NTVSAMGPFK GLKEVRRVVE DCMKNIHPIY HIKELMIKRE LAKRPELANE DWSRFLPMFK KRNVARKKPK KI RNVEKKV YTPFPPAQLP RKVDLEIESG EYFLSKREKQ MKKLNEQKEK QMEREIERQE ERAKDFIAPE EEAYKPNQN UniProtKB: KRR1 small subunit processome component |
+Macromolecule #45: Nop56
| Macromolecule | Name: Nop56 / type: protein_or_peptide / ID: 45 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 56.394863 KDa |
| Sequence | String: MAPIEYLLFE EPTGYAVFKV KLQQDDIGSR LKEVQEQIND FGAFTKLIEL VSFAPFKGAA EALENANDIS EGLVSESLKA ILDLNLPKA SSKKKNITLA ISDKNLGPSI KEEFPYVDCI SNELAQDLIR GVRLHGEKLF KGLQSGDLER AQLGLGHAYS R AKVKFSVQ ...String: MAPIEYLLFE EPTGYAVFKV KLQQDDIGSR LKEVQEQIND FGAFTKLIEL VSFAPFKGAA EALENANDIS EGLVSESLKA ILDLNLPKA SSKKKNITLA ISDKNLGPSI KEEFPYVDCI SNELAQDLIR GVRLHGEKLF KGLQSGDLER AQLGLGHAYS R AKVKFSVQ KNDNHIIQAI ALLDQLDKDI NTFAMRVKEW YGWHFPELAK LVPDNYTFAK LVLFIKDKA(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)NAR ISMGQDISET DMENVCVFAQ RVASLADYRR QLYDYLC EK MHTVAPNLSE LIGEVIGARL ISHAGSLTNL SKQAASTVQI LGAEKALFRA LKTKGNTPKY GLIYHSGFIS KASAKNKG R ISRYLANKCS MASRIDNYSE EPSNVFGSVL KKQVEQRLEF YNTGKPTLKN ELAIQEAMEL YNKDKPAAEV EETKEKESS KKRKLEDDDE EKKEKKEKKS KKEKKEKKEK KDKKEKKDKK EKKDKKKKSK D UniProtKB: Nucleolar protein 56, Nucleolar protein 56, Nucleolar protein 56 |
+Macromolecule #46: Nop58
| Macromolecule | Name: Nop58 / type: protein_or_peptide / ID: 46 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 53.81452 KDa |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) ...String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) DNDLSKMSLG LAHSIGRHKL KFSADKVDVM IIQAIALLDD LDKELNTYAM RCKEWYGWHF PELAKIVTD SVAYARIILT MGIRSKASET DLSEILPEEI EERVKTAAEV SMGTEITQTD LDNINALAEQ IVEFAAYREQ L SNYLSARM KAIAPNLTQL VGELVGARLI AHSGSLISLA KSPASTIQIL GAEKALFRAL KTKHDTPKYG LLYHASLVGQ AT GKNKGKI ARVLAAKAAV SLRYDALAED RDDSGDIGLE SRAKVENRLS QLEGRDLRTT PKVVREAKKV EMTEARAYNA DAD TAKAAS DSESDSDDEE EEKKEKKEKK RKRDDDEDSK DSKKAKKEKK DKKEKKEKKE KKEKKEKKEK KEKKSKKEKK EKK UniProtKB: Nucleolar protein 58 |
+Macromolecule #47: Nop1
| Macromolecule | Name: Nop1 / type: protein_or_peptide / ID: 47 / Number of copies: 2 / Enantiomer: LEVO EC number: Transferases; Transferring one-carbon groups; Methyltransferases |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 34.525418 KDa |
| Sequence | String: MSFRPGSRGG SRGGSRGGFG GRGGSRGGAR GGSRGGFGGR GGSRGGARGG SRGGFGGRGG SRGGARGGSR GGRGGAAGGA RGGAKVVIE PHRHAGVYIA RGKEDLLVTK NMAPGESVYG EKRISVEEPS KEDGVPPTKV EYRVWNPFRS KLAAGIMGGL D ELFIAPGK ...String: MSFRPGSRGG SRGGSRGGFG GRGGSRGGAR GGSRGGFGGR GGSRGGARGG SRGGFGGRGG SRGGARGGSR GGRGGAAGGA RGGAKVVIE PHRHAGVYIA RGKEDLLVTK NMAPGESVYG EKRISVEEPS KEDGVPPTKV EYRVWNPFRS KLAAGIMGGL D ELFIAPGK KVLYLGAASG TSVSHVSDVV GPEGVVYAVE FSHRPGRELI SMAKKRPNII PIIEDARHPQ KYRMLIGMVD CV FADVAQP DQARIIALNS HMFLKDQGGV VISIKANCID STVDAETVFA REVQKLREER IKPLEQLTLE PYERDHCIVV GRY MRSGLK K UniProtKB: rRNA 2'-O-methyltransferase fibrillarin |
+Macromolecule #48: Snu13
| Macromolecule | Name: Snu13 / type: protein_or_peptide / ID: 48 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.582855 KDa |
| Sequence | String: MSAPNPKAFP LADAALTQQI LDVVQQAANL RQLKKGANEA TKTLNRGISE FIIMAADCEP IEILLHLPLL CEDKNVPYVF VPSRVALGR ACGVSRPVIA ASITTNDASA IKTQIYAVKD KIETLLI UniProtKB: 13 kDa ribonucleoprotein-associated protein |
+Macromolecule #49: Rrp9
| Macromolecule | Name: Rrp9 / type: protein_or_peptide / ID: 49 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 65.146969 KDa |
| Sequence | String: MSDVTQQKKR KRSKGEVNPS KPTVDEEITD PSSNEDEQLE VSDEEDALES EEEFEGENPA DKRRRLAKQY LENLKSEAND ILTDNRNAE EKDLNNLKER TIDEYNNFDA GDLDKDIIAS RLKEDVAEQQ GRVFRYFGDK LLISEAKQSF TRVGENNLTC I SCFQPVLN ...String: MSDVTQQKKR KRSKGEVNPS KPTVDEEITD PSSNEDEQLE VSDEEDALES EEEFEGENPA DKRRRLAKQY LENLKSEAND ILTDNRNAE EKDLNNLKER TIDEYNNFDA GDLDKDIIAS RLKEDVAEQQ GRVFRYFGDK LLISEAKQSF TRVGENNLTC I SCFQPVLN KYTFEESSNG DKNKGRLFAY TVSKDLQLTK YDITDFSKRP KKLKYAKGGA KYIPTSKHEY ENTTEGHYDE IL TVAASPD GKYVVTGGRD RKLIVWSTES LSPVKVIPTK DRRGEVLSLA FRKNSDQLYA SCADFKIRTY SINQFSQLEI LYG HHDIVE DISALAMERC VTVGARDRTA MLWKIPDETR LTFRGGDEPQ KLLRRWMKEN AKEGEDGEVK YPDESEAPLF FCEG SIDVV SMVDDFHFIT GSDNGNICLW SLAKKKPIFT ERIAHGILPE PSFNDISGET DEELRKRQLQ GKKLLQPFWI TSLYA IPYS NVFISGSWSG SLKVWKISDN LRSFELLGEL SGAKGVVTKI QVVESGKHGK EKFRILASIA KEHRLGRWIA NVSGAR NGI YSAVIDQTGF UniProtKB: Ribosomal RNA-processing protein 9 |
+Macromolecule #50: Rcl1
| Macromolecule | Name: Rcl1 / type: protein_or_peptide / ID: 50 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 40.220559 KDa |
| Sequence | String: MSSSAPKYTT FQGSQNFRLR IVLATLSGKP IKIEKIRSGD LNPGLKDYEV SFLRLIESVT NGSVIEISYT GTTVIYRPGI IVGGASTHI CPSSKPVGYF VEPMLYLAPF SKKKFSILFK GITASHNDAG IEAIKWGLMP VMEKFGVREC ALHTLKRGSP P LGGGEVHL ...String: MSSSAPKYTT FQGSQNFRLR IVLATLSGKP IKIEKIRSGD LNPGLKDYEV SFLRLIESVT NGSVIEISYT GTTVIYRPGI IVGGASTHI CPSSKPVGYF VEPMLYLAPF SKKKFSILFK GITASHNDAG IEAIKWGLMP VMEKFGVREC ALHTLKRGSP P LGGGEVHL VVDSLIAQPI TMHEIDRPII SSITGVAYST RVSPSLVNRM IDGAKKVLKN LQCEVNITAD VWRGENSGKS PG WGITLVA QSKQKGWSYF AEDIGDAGSI PEELGEKVAC QLLEEISKSA AVGRNQLPLA IVYMVIGKED IGRLRINKEQ IDE RFIILL RDIKKIFNTE VFLKPVDEAD NEDMIATIKG IGFTNTSKKI A UniProtKB: rRNA processing protein RCL1 |
+Macromolecule #51: Bms1
| Macromolecule | Name: Bms1 / type: protein_or_peptide / ID: 51 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 135.152938 KDa |
| Sequence | String: MEQSNKQHRK AKEKNTAKKK LHTQGHNAKA FAVAAPGKMA RTMQRSSDVN ERKLHVPMVD RTPEDDPPPF IVAVVGPPGT GKTTLIRSL VRRMTKSTLN DIQGPITVVS GKHRRLTFLE CPADDLNAMI DIAKIADLVL LLIDGNFGFE METMEFLNIA Q HHGMPRVL ...String: MEQSNKQHRK AKEKNTAKKK LHTQGHNAKA FAVAAPGKMA RTMQRSSDVN ERKLHVPMVD RTPEDDPPPF IVAVVGPPGT GKTTLIRSL VRRMTKSTLN DIQGPITVVS GKHRRLTFLE CPADDLNAMI DIAKIADLVL LLIDGNFGFE METMEFLNIA Q HHGMPRVL GVATHLDLFK SQSTLRASKK RLKHRFWTEV YQGAKLFYLS GVINGRYPDR EILNLSRFIS VMKFRPLKWR NE HPYMLAD RFTDLTHPEL IETQGLQIDR KVAIYGYLHG TPLPSAPGTR VHIAGVGDFS VAQIEKLPDP CPTPFYQQKL DDF EREKMK EEAKANGEIT TASTTRRRKR LDDKDKLIYA PMSDVGGVLM DKDAVYIDIG KKNEEPSFVP G(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)FSNGT ELHEV ADHE GMDVESGEES IEDDEGKSKG RTSLRKPRIY GKPVQEEDAD IDNLPSDEEP YTNDDDVQDS EPRMVEIDFN NTGEQG AEK LALETDSEFE ESEDEFSWER TAANKLKKTE SKKRTWNIGK LIYMDNISPE ECIRRWRGED DDSKDESDIE EDVDDDF FR KKDGTVTKEG NKDHAVDLEK FVPYFDTFEK LAKKWKSVDA IKERFLGAGI LGNDNKTKSD SNEGGEELYG DFEDLEDG N PSEQAEDNSD KESEDEDENE DTNGDDDNSF TNFDAEEKKD LTMEQEREMN AAKKEKLRAQ FEIEEGENFK EDDENNEYD TWYELQKAKI SKQLEINNIE YQEMTPEQRQ RIEGFKAGSY VRIVFEKVPM EFVKNFNPKF PIVMGGLLPT EIKFGIVKAR LRRHRWHKK ILKTNDPLVL SLGWRRFQTL PIYTTTDSRT RTRMLKYTPE HTYCNAAFYG PLCSPNTPFC GVQIVANSDT G NGFRIAAT GIVEEIDVNI EIVKKLKLVG FPYKIFKNTA FIKDMFSSAM EVARFEGAQI KTVSGIRGEI KRALSKPEGH YR AAFEDKI LMSDIVILRS WYPVRVKKFY NPVTSLLLKE KTEWKGLRLT GQIRAAMNLE TPSNPDSAYH KIERVERHFN GLK VPKAVQ KELPFKSQIH QMKPQKKKTY MAKRAVVLGG DEKKARSFIQ KVLTISKAKD SKRKEQKASQ RKERLKKLAK MEEE KSQRD KEKKKEYFAQ NGKRTTMGGD DESRPRKMRR UniProtKB: Ribosome biogenesis protein BMS1, Ribosome biogenesis protein BMS1 |
+Macromolecule #52: Emg1
| Macromolecule | Name: Emg1 / type: protein_or_peptide / ID: 52 / Number of copies: 2 / Enantiomer: LEVO EC number: rRNA small subunit pseudouridine methyltransferase Nep1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 27.936461 KDa |
| Sequence | String: MVEDSRVRDA LKGGDQKALP ASLVPQAPPV LTSKDKITKR MIVVLAMASL ETHKISSNGP GGDKYVLLNC DDHQGLLKKM GRDISEARP DITHQCLLTL LDSPINKAGK LQVYIQTSRG ILIEVNPTVR IPRTFKRFSG LMVQLLHKLS IRSVNSEEKL L KVIKNPIT ...String: MVEDSRVRDA LKGGDQKALP ASLVPQAPPV LTSKDKITKR MIVVLAMASL ETHKISSNGP GGDKYVLLNC DDHQGLLKKM GRDISEARP DITHQCLLTL LDSPINKAGK LQVYIQTSRG ILIEVNPTVR IPRTFKRFSG LMVQLLHKLS IRSVNSEEKL L KVIKNPIT DHLPTKCRKV TLSFDAPVIR VQDYIEKLDD DESICVFVGA MARGKDNFAD EYVDEKVGLS NYPLSASVAC SK FCHGAED AWNIL UniProtKB: Ribosomal RNA small subunit methyltransferase NEP1 |
+Macromolecule #53: Utp24
| Macromolecule | Name: Utp24 / type: protein_or_peptide / ID: 53 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 21.650729 KDa |
| Sequence | String: MGKAKKTRKF GLVKRTLNTK KDQRLKKNQE NIKTKEDPEL TRNIPQVSSA LFFQYNQAIK PPYQVLIDTN FINFSIQKKV DIVRGMMDC LLAKCNPLIT DCVMAELEKL GPKYRIALKL ARDPRIKRLS CSHKGTYADD CLVHRVLQHK CYIVATNDAG L KQRIRKIP GIPLMSVGGH AYVIEKLPDV F UniProtKB: rRNA-processing protein FCF1 |
+Macromolecule #54: Imp4
| Macromolecule | Name: Imp4 / type: protein_or_peptide / ID: 54 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 33.536168 KDa |
| Sequence | String: MLRRQARERR EYLYRKAQEL QDSQLQQKRQ IIKQALAQGK PLPKELAEDE SLQKDFRYDQ SLKESEEADD LQVDDEYAAT SGIMDPRII VTTSRDPSTR LSQFAKEIKL LFPNAVRLNR GNYVMPNLVD ACKKSGTTDL VVLHEHRGVP TSLTISHFPH G PTAQFSLH ...String: MLRRQARERR EYLYRKAQEL QDSQLQQKRQ IIKQALAQGK PLPKELAEDE SLQKDFRYDQ SLKESEEADD LQVDDEYAAT SGIMDPRII VTTSRDPSTR LSQFAKEIKL LFPNAVRLNR GNYVMPNLVD ACKKSGTTDL VVLHEHRGVP TSLTISHFPH G PTAQFSLH NVVMRHDIIN AGNQSEVNPH LIFDNFTTAL GKRVVCILKH LFNAGPKKDS ERVITFANRG DFISVRQHVY VR TREGVEI AEVGPRFEMR LFELRLGTLE NKDADVEWQL RRFIRTANKK DYL UniProtKB: U3 small nucleolar ribonucleoprotein protein IMP4 |
+Macromolecule #55: Utp30
| Macromolecule | Name: Utp30 / type: protein_or_peptide / ID: 55 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 31.668939 KDa |
| Sequence | String: MVESNDIIKS GLAEKALKAL ILQCEENPSL KNDKDIHIII NTGKKMGINR DNIPRIIPLT KYKLFKPRDL NILLITKDPS ALYRETLTK DEHTSELFKE IISVKNLRRR FKGSKLTQLY KDFDLVVADY RVHHLLPEVL GSRFYHGSKK LPYMIRMSKE V KLKRQQMV ...String: MVESNDIIKS GLAEKALKAL ILQCEENPSL KNDKDIHIII NTGKKMGINR DNIPRIIPLT KYKLFKPRDL NILLITKDPS ALYRETLTK DEHTSELFKE IISVKNLRRR FKGSKLTQLY KDFDLVVADY RVHHLLPEVL GSRFYHGSKK LPYMIRMSKE V KLKRQQMV EKCDPIYVRA QLRSICKNTS YIPNNDNCLS VRVGYIQKHS IPEILQNIQD TINFLTDKSK RPQGGVIKGG II SIFVKTS NSTSLPIYQF SEARENQKNE DLSDIKL UniProtKB: Ribosome biogenesis protein UTP30 |
+Macromolecule #56: Pno1
| Macromolecule | Name: Pno1 / type: protein_or_peptide / ID: 56 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 30.380623 KDa |
| Sequence | String: MVAPTALKKA TVTPVSGQDG GSSRIIGINN TESIDEDDDD DVLLDDSDNN TAKEEVEGEE GSRKTHESKT VVVDDQGKPR FTSASKTQG NKIKFESRKI MVPPHRMTPL RNSWTKIYPP LVEHLKLQVR MNLKTKSVEL RTNPKFTTDP GALQKGADFI K AFTLGFDL ...String: MVAPTALKKA TVTPVSGQDG GSSRIIGINN TESIDEDDDD DVLLDDSDNN TAKEEVEGEE GSRKTHESKT VVVDDQGKPR FTSASKTQG NKIKFESRKI MVPPHRMTPL RNSWTKIYPP LVEHLKLQVR MNLKTKSVEL RTNPKFTTDP GALQKGADFI K AFTLGFDL DDSIALLRLD DLYIETFEVK DVKTLTGDHL SRAIGRIAGK DGKTKFAIEN ATRTRIVLAD SKIHILGGFT HI RMARESV VSLILGSPPG KVYGNLRTVA SRLKERY UniProtKB: Pre-rRNA-processing protein PNO1 |
+Macromolecule #57: Utp20
| Macromolecule | Name: Utp20 / type: protein_or_peptide / ID: 57 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 83.59018 KDa |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) ...String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK) |
+Macromolecule #58: Fcf2
| Macromolecule | Name: Fcf2 / type: protein_or_peptide / ID: 58 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 25.68924 KDa |
| Sequence | String: MDQSVEDLFG ALRDASASLE VKNSAKEQVS LQQEDVLQIG NNDDEVEIES KFQEIETNLK KLPKLETGFD ALANKKKKKN VLPSVETED KRKPNKSDKN DNDWFTLPKP DDNMRREVQR DLLLIKHRAA LDPKRHYKKQ RWEVPERFAI GTIIEDKSEF Y SSRMNRKE ...String: MDQSVEDLFG ALRDASASLE VKNSAKEQVS LQQEDVLQIG NNDDEVEIES KFQEIETNLK KLPKLETGFD ALANKKKKKN VLPSVETED KRKPNKSDKN DNDWFTLPKP DDNMRREVQR DLLLIKHRAA LDPKRHYKKQ RWEVPERFAI GTIIEDKSEF Y SSRMNRKE RKSTILETLM GDEASNKYFK RKYNEIQEKS TSGRKAHYKK MKEMRKKRR UniProtKB: rRNA-processing protein FCF2 |
+Macromolecule #59: rpS23_uS12
| Macromolecule | Name: rpS23_uS12 / type: protein_or_peptide / ID: 59 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 16.073896 KDa |
| Sequence | String: MGKGKPRGLN SARKLRVHRR NNRWAENNYK KRLLGTAFKS SPFGGSSHAK GIVLEKLGIE SKQPNSAIRK CVRVQLIKNG KKVTAFVPN DGCLNFVDEN DEVLLAGFGR KGKAKGDIPG VRFKVVKVSG VSLLALWKEK KEKPRS UniProtKB: Small ribosomal subunit protein uS12A |
+Macromolecule #60: Utp14
| Macromolecule | Name: Utp14 / type: protein_or_peptide / ID: 60 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 101.101461 KDa |
| Sequence | String: MAKKKSKSRS KSSRRVLDAL QLAEREINGE FDNSSDNDKR HDARRNGTVV NLLKRSKGDT NSDEDDIDSE SFEDEELNSD EALGSDDDY DILNSKFSQT IRDKKENANY QEEEDEGGYT SIDEEDLMPL SQVWDMDEKT AQSNGNDDED ASPQLKLQDT D ISSESSSS ...String: MAKKKSKSRS KSSRRVLDAL QLAEREINGE FDNSSDNDKR HDARRNGTVV NLLKRSKGDT NSDEDDIDSE SFEDEELNSD EALGSDDDY DILNSKFSQT IRDKKENANY QEEEDEGGYT SIDEEDLMPL SQVWDMDEKT AQSNGNDDED ASPQLKLQDT D ISSESSSS EESESESEDD EEEEDPFDEI SEDEEDIELN TITSKLIDET KSKAPKRLDT YGSGEANEYV LPSANAASGA SG KLSLTDM MNVIDDRQVI ENANLLKGKS STYEVPLPQR IQQRHDRKAA YEISRQEVSK WNDIVQQNRR ADHLIFPLNK PTE HNHASA FTRTQDVPQT ELQEKVDQVL QESN(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)T YRKIKKKELM KNRELAAVSS DEDNEDHDI ARAKERMTLK HKTNSKWAKD MIKHGMTNDA ETREEMEEML RQGERLKAKM LDRNSDDEED GRVQTLSDVE NEEKENIDSE ALKSKLGKT GVMNMAFMKN GEAREREANK ETLRQLRAVE NGDDIKLFES DEEETNGENI QINKGRRIYT PGSLESNKDM N ELNDHTRK ENKVDESRSL ENRLRAKNSG QSKNARTNAE GAIIVEEESD GEPLQDGQNN QQDEEAKDVN PWLANESDEE HT VKKQSSK VNVIDKDSSK NVKAMNKMEK AELKQKKKKK GKSNDDEDLL LTADDSTRLK IVDPYGGSDD EQGDNVFMFK QQD VIAEAF AGDDVVAEFQ EEKKRVIDDE DDKEVDTTLP GWGEWAGAGS KPKNKKRKFI KKVKGVVNKD KRRDKNLQNV IINE KVNKK NLKYQSSAVP FPFENREQYE RSLRMPIGQE WTSRASHQEL IKPRIMTKPG QVIDPLKAPF K UniProtKB: U3 small nucleolar RNA-associated protein 14, U3 small nucleolar RNA-associated protein 14 |
+Macromolecule #61: Nop14
| Macromolecule | Name: Nop14 / type: protein_or_peptide / ID: 61 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 85.993961 KDa |
| Sequence | String: MAGSQLKNLK AALKARGLTG QTNVKSKNKK NSKRQAKEYD REEKKKAIAE IREEFNPFEI KAARNKRRDG LPSKTADRIA VGKPGISKQ IGEEQRKRAF EARKMMKNKR GGVIDKRFGE RDKLLTEEEK MLERFTRERQ SQSKRNANLF NLEDDEDDGD M FGDGLTHL ...String: MAGSQLKNLK AALKARGLTG QTNVKSKNKK NSKRQAKEYD REEKKKAIAE IREEFNPFEI KAARNKRRDG LPSKTADRIA VGKPGISKQ IGEEQRKRAF EARKMMKNKR GGVIDKRFGE RDKLLTEEEK MLERFTRERQ SQSKRNANLF NLEDDEDDGD M FGDGLTHL GQSLSLEDEL ANDEEDFLAS KRFNEDDAEL QQ(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) KEKLG KFTAVLLRHI IFLSNQNYLK NVQSFKRTQN ALISILKSLS EKYNRELSEE CRDYINEMQA RYKKNHFDAL SNGDL VFFS IIGILFSTSD QYHLVITPAL ILMSQFLEQI KFNSLKRIAF GAVLVRIVSQ YQRISKRYIP EVVYFFQKIL LTFIVE KEN QEKPLDFENI RLDSYELGLP LDVDFTKKRS TIIPLHTLST MDTEAHPVDQ CVSVLLNVME SLDATISTVW KSLPAFN EI ILPIQQLLSA YTSKYSDFEK PRNILNKVEK LTKFTEHIPL ALQNHKPVSI PTHAPKYEEN FNPDKKSYDP DRTRSEIN K MKAQLKKERK FTMKEIRKDA KFEARQRIEE KNKESSDYHA KMAHIVNTIN TEEGAEKNKY ERERKLRGGK K UniProtKB: Nucleolar complex protein 14, Nucleolar complex protein 14 |
+Macromolecule #62: Noc4
| Macromolecule | Name: Noc4 / type: protein_or_peptide / ID: 62 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 58.369125 KDa |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) ...String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)FRKL IIALWSSNMG EIEDVKSSGA SENLIIVEFT EKYYTKFADI QYYFQSEFNQ LLEDPAYQDL L LKNVGKWL ALVNHDKHCS SVDADLEIFV PNPPQAIENE SKFKSNFEKN WLSLLNGQLS LQQYKSILLI LHKRIIPHFH TP TKLMDFL TDSYNLQSSN KNAGVVPILA LNGLFELMKR FNLEYPNFYM KLYQIINPDL MHVKYRARFF RLMDVFLSST HLS AHLVAS FIKKLARLTL ESPPSAIVTV IPFIYNLIRK HPNCMIMLHN PAFISNPFQT PDQVANLKTL KENYVDPFDV HESD PELTH ALDSSLWELA SLMEHYHPNV ATLAKIFAQP FKKLSYNMED FLDWNYDSLL NAESSR(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) UniProtKB: Nucleolar complex protein 4 |
+Macromolecule #63: Rrt14
| Macromolecule | Name: Rrt14 / type: protein_or_peptide / ID: 63 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 21.036877 KDa |
| Sequence | String: MSSSLSQTSK YQATSVVNGL LSNLLPGVPK IRANNGKTSV NNGSKAQLID RNLKKRVQLQ NRDVHKIKKK CKLVKKKKVK KHKLDKEQL EQLAKHQVLK K(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK) ...String: MSSSLSQTSK YQATSVVNGL LSNLLPGVPK IRANNGKTSV NNGSKAQLID RNLKKRVQLQ NRDVHKIKKK CKLVKKKKVK KHKLDKEQL EQLAKHQVLK K(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)FVKDHRY PGLTPGLAPV GLSDEEDSSE ED UniProtKB: Regulator of rDNA transcription 14, Regulator of rDNA transcription 14 |
+Macromolecule #64: Unassigned peptides
| Macromolecule | Name: Unassigned peptides / type: protein_or_peptide / ID: 64 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 8.783818 KDa |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) ...String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) |
+Macromolecule #65: Utp11
| Macromolecule | Name: Utp11 / type: protein_or_peptide / ID: 65 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 29.806348 KDa |
| Sequence | String: MAKLVHDVQK KQHRERSQLT SRSRYGFLEK HKDYVKRAQD FHRKQSTLKV LREKAKERNP DEYYHAMHSR KTDAKGLLIS SRHGDEEDE SLSMDQVKLL KTQDSNYVRT LRQIELKKLE KGAKQLMFKS SGNHTIFVDS REKMNEFTPE KFFNTTSEMV N RSENRLTK ...String: MAKLVHDVQK KQHRERSQLT SRSRYGFLEK HKDYVKRAQD FHRKQSTLKV LREKAKERNP DEYYHAMHSR KTDAKGLLIS SRHGDEEDE SLSMDQVKLL KTQDSNYVRT LRQIELKKLE KGAKQLMFKS SGNHTIFVDS REKMNEFTPE KFFNTTSEMV N RSENRLTK DQLAQDISNN RNASSIMPKE SLDKKKLKKF KQVKQHLQRE TQLKQVQQRM DAQRELLKKG SKKKIVDSSG KI SFKWKKQ RKR UniProtKB: U3 small nucleolar RNA-associated protein 11 |
+Macromolecule #66: Enp1
| Macromolecule | Name: Enp1 / type: protein_or_peptide / ID: 66 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 55.207422 KDa |
| Sequence | String: MARASSTKAR KQRHDPLLKD LDAAQGTLKK INKKKLAQND AANHDAANEE DGYIDSKASR KILQLAKEQQ DEIEGEELAE SERNKQFEA RFTTMSYDDE DEDEDEDEEA FGEDISDFEP EGDYKEEEEI VEIDEEDAAM FEQYFKKSDD FNSLSGSYNL A DKIMASIR ...String: MARASSTKAR KQRHDPLLKD LDAAQGTLKK INKKKLAQND AANHDAANEE DGYIDSKASR KILQLAKEQQ DEIEGEELAE SERNKQFEA RFTTMSYDDE DEDEDEDEEA FGEDISDFEP EGDYKEEEEI VEIDEEDAAM FEQYFKKSDD FNSLSGSYNL A DKIMASIR EKESQVEDMQ DDEPLANEQN TSRGNISSGL KSGEGVALPE KVIKAYTTVG SILKTWTHGK LPKLFKVIPS LR NWQDVIY VTNPEEWSPH VVYEATKLFV SNLTAKESQK FINLILLERF RDNIETSEDH SLNYHIYRAV KKSLYKPSAF FKG FLFPLV ETGCNVREAT IAGSVLAKVS VPALHSSAAL SYLLRLPFSP PTTVFIKILL DKKYALPYQT VDDCVYYFMR FRIL DDGSN GEDATRVLPV IWHKAFLTFA QRYKNDITQD QRDFLLETVR QRGHKDIGPE IRRELLAGAS REFVDPQEAN DDLMI DVN UniProtKB: Essential nuclear protein 1 |
+Macromolecule #67: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 67 / Number of copies: 1 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.15 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.7 Component:
| ||||||||||||||||||
| Grid | Model: Ted Pella Inc. / Material: COPPER / Mesh: 400 / Support film - #0 - Film type ID: 1 / Support film - #0 - Material: CARBON / Support film - #0 - topology: LACEY / Support film - #1 - Film type ID: 2 / Support film - #1 - Material: CARBON / Support film - #1 - topology: CONTINUOUS / Support film - #1 - Film thickness: 2 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR / Details: 50 mA | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 295 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Number grids imaged: 4 / Number real images: 10029 / Average exposure time: 0.25 sec. / Average electron dose: 1.56 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.6 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 22500 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: OTHER / Overall B value: 338.9 |
|---|---|
| Output model |  PDB-5wlc: |
+ About Yorodumi
About Yorodumi
-News
-Feb 9, 2022. New format data for meta-information of EMDB entries
New format data for meta-information of EMDB entries
- Version 3 of the EMDB header file is now the official format.
- The previous official version 1.9 will be removed from the archive.
Related info.: EMDB header
EMDB header
External links: wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
-Aug 12, 2020. Covid-19 info
Covid-19 info

URL: https://pdbj.org/emnavi/covid19.php
New page: Covid-19 featured information page in EM Navigator.
Related info.: Covid-19 info /
Covid-19 info /  Mar 5, 2020. Novel coronavirus structure data
Mar 5, 2020. Novel coronavirus structure data
+Mar 5, 2020. Novel coronavirus structure data
Novel coronavirus structure data
- International Committee on Taxonomy of Viruses (ICTV) defined the short name of the 2019 coronavirus as "SARS-CoV-2".
- In the structure databanks used in Yorodumi, some data are registered as the other names, "COVID-19 virus" and "2019-nCoV". Here are the details of the virus and the list of structure data.
Related info.: Yorodumi Speices /
Yorodumi Speices /  Aug 12, 2020. Covid-19 info
Aug 12, 2020. Covid-19 info
External links: COVID-19 featured content - PDBj /
COVID-19 featured content - PDBj /  Molecule of the Month (242):Coronavirus Proteases
Molecule of the Month (242):Coronavirus Proteases
+Jan 31, 2019. EMDB accession codes are about to change! (news from PDBe EMDB page)
EMDB accession codes are about to change! (news from PDBe EMDB page)
- The allocation of 4 digits for EMDB accession codes will soon come to an end. Whilst these codes will remain in use, new EMDB accession codes will include an additional digit and will expand incrementally as the available range of codes is exhausted. The current 4-digit format prefixed with “EMD-” (i.e. EMD-XXXX) will advance to a 5-digit format (i.e. EMD-XXXXX), and so on. It is currently estimated that the 4-digit codes will be depleted around Spring 2019, at which point the 5-digit format will come into force.
- The EM Navigator/Yorodumi systems omit the EMD- prefix.
Related info.: Q: What is EMD? /
Q: What is EMD? /  ID/Accession-code notation in Yorodumi/EM Navigator
ID/Accession-code notation in Yorodumi/EM Navigator
External links: EMDB Accession Codes are Changing Soon! /
EMDB Accession Codes are Changing Soon! /  Contact to PDBj
Contact to PDBj
+Jul 12, 2017. Major update of PDB
Major update of PDB
- wwPDB released updated PDB data conforming to the new PDBx/mmCIF dictionary.
- This is a major update changing the version number from 4 to 5, and with Remediation, in which all the entries are updated.
- In this update, many items about electron microscopy experimental information are reorganized (e.g. em_software).
- Now, EM Navigator and Yorodumi are based on the updated data.
External links: wwPDB Remediation /
wwPDB Remediation /  Enriched Model Files Conforming to OneDep Data Standards Now Available in the PDB FTP Archive
Enriched Model Files Conforming to OneDep Data Standards Now Available in the PDB FTP Archive
-Yorodumi
Thousand views of thousand structures
- Yorodumi is a browser for structure data from EMDB, PDB, SASBDB, etc.
- This page is also the successor to EM Navigator detail page, and also detail information page/front-end page for Omokage search.
- The word "yorodu" (or yorozu) is an old Japanese word meaning "ten thousand". "mi" (miru) is to see.
Related info.: EMDB /
EMDB /  PDB /
PDB /  SASBDB /
SASBDB /  Comparison of 3 databanks /
Comparison of 3 databanks /  Yorodumi Search /
Yorodumi Search /  Aug 31, 2016. New EM Navigator & Yorodumi /
Aug 31, 2016. New EM Navigator & Yorodumi /  Yorodumi Papers /
Yorodumi Papers /  Jmol/JSmol /
Jmol/JSmol /  Function and homology information /
Function and homology information /  Changes in new EM Navigator and Yorodumi
Changes in new EM Navigator and Yorodumi
 Movie
Movie Controller
Controller





























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)