+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6665 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
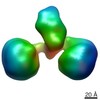
| Title | The hexamer of full-length Cbln1 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 13.0 Å | |||||||||
 Authors Authors | Zhong C / Li G / Zhang H / Cao L / He Y / Ding J | |||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2017 Journal: Cell Rep / Year: 2017Title: Cbln1 and Cbln4 Are Structurally Similar but Differ in GluD2 Binding Interactions. Authors: Chen Zhong / Jinlong Shen / Huibing Zhang / Guangyi Li / Senlin Shen / Fang Wang / Kuan Hu / Longxing Cao / Yongning He / Jianping Ding /  Abstract: Unlike cerebellin 1 (Cbln1), which bridges neurexin (Nrxn) receptors and δ-type glutamate receptors in a trans-synaptic triad, Cbln4 was reported to have no or weak binding for the receptors ...Unlike cerebellin 1 (Cbln1), which bridges neurexin (Nrxn) receptors and δ-type glutamate receptors in a trans-synaptic triad, Cbln4 was reported to have no or weak binding for the receptors despite sharing ∼70% sequence identity with Cbln1. Here, we report crystal structures of the homotrimers of the C1q domain of Cbln1 and Cbln4 at 2.2 and 2.3 Å resolution, respectively. Comparison of the structures suggests that the difference between Cbln1 and Cbln4 in GluD2 binding might be because of their sequence and structural divergence in loop CD. Surprisingly, we show that Cbln4 binds to Nrxn1β and forms a stable complex with the laminin, nectin, sex-hormone binding globulin (LNS) domain of Nrxn1β. Furthermore, the negative-stain electron microscopy reconstruction of hexameric full-length Cbln1 at 13 Å resolution and that of the Cbln4/Nrxn1β complex at 19 Å resolution suggest that Nrxn1β binds to the N-terminal region of Cbln4, probably through strand β10 of the S4 insert. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6665.map.gz emd_6665.map.gz | 868.1 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6665-v30.xml emd-6665-v30.xml emd-6665.xml emd-6665.xml | 8.4 KB 8.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_6665.png emd_6665.png | 35.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6665 http://ftp.pdbj.org/pub/emdb/structures/EMD-6665 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6665 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6665 | HTTPS FTP |
-Validation report
| Summary document |  emd_6665_validation.pdf.gz emd_6665_validation.pdf.gz | 78.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_6665_full_validation.pdf.gz emd_6665_full_validation.pdf.gz | 77.7 KB | Display | |
| Data in XML |  emd_6665_validation.xml.gz emd_6665_validation.xml.gz | 494 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6665 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6665 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6665 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6665 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_6665.map.gz / Format: CCP4 / Size: 1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6665.map.gz / Format: CCP4 / Size: 1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.96 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Cbln1
| Entire | Name: Cbln1 |
|---|---|
| Components |
|
-Supramolecule #1: Cbln1
| Supramolecule | Name: Cbln1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant plasmid: pCDH Homo sapiens (human) / Recombinant plasmid: pCDH |
-Macromolecule #1: Cbln1
| Macromolecule | Name: Cbln1 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GTHHHHHHHH GSQNETEPIV LEGKCLVVCD SNPTSDPTGT ALGISVRSGS AKVAFSAIRS TNHEPSEMSN RTMIIYFDQV LVNIGNNFDS ERSTFIAPRK GIYSFNFHVV KVYNRQTIQV SLMLNGWPVI SAFAGDQDVT REAASNGVLI QMEKGDRAYL KLERGNLMGG WKYSTFSGFL VFPL |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Staining | Type: NEGATIVE / Material: 0.75% uranyl formate |
| Vitrification | Cryogen name: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Image recording | Film or detector model: FEI EAGLE (4k x 4k) / Average electron dose: 20.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 13.0 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 41994 |
|---|---|
| Initial angle assignment | Type: NOT APPLICABLE |
| Final angle assignment | Type: NOT APPLICABLE |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















