+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6495 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Electron microscopy of apo human XPC complex | |||||||||
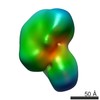
 Map data Map data | EMAN2 reconstruction of apo human XPC complex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | transcription / DNA repair / stem cells | |||||||||
| Function / homology |  Function and homology information Function and homology informationheteroduplex DNA loop binding / pyrimidine dimer repair by nucleotide-excision repair / nucleotide-excision repair factor 2 complex / XPC complex / 9+2 motile cilium / nucleotide-excision repair complex / photoreceptor connecting cilium / DNA damage sensor activity / regulation of proteasomal ubiquitin-dependent protein catabolic process / heterotrimeric G-protein binding ...heteroduplex DNA loop binding / pyrimidine dimer repair by nucleotide-excision repair / nucleotide-excision repair factor 2 complex / XPC complex / 9+2 motile cilium / nucleotide-excision repair complex / photoreceptor connecting cilium / DNA damage sensor activity / regulation of proteasomal ubiquitin-dependent protein catabolic process / heterotrimeric G-protein binding / transcription export complex 2 / response to auditory stimulus / nuclear pore nuclear basket / bubble DNA binding / UV-damage excision repair / cellular response to interleukin-7 / response to UV-B / mitotic intra-S DNA damage checkpoint signaling / regulation of mitotic cell cycle phase transition / proteasome binding / centriole replication / site of DNA damage / glial cell projection / polyubiquitin modification-dependent protein binding / mRNA transport / mismatch repair / SUMOylation of DNA damage response and repair proteins / centriole / Loss of Nlp from mitotic centrosomes / Loss of proteins required for interphase microtubule organization from the centrosome / Recruitment of mitotic centrosome proteins and complexes / Recruitment of NuMA to mitotic centrosomes / Anchoring of the basal body to the plasma membrane / proteasome complex / AURKA Activation by TPX2 / Josephin domain DUBs / ubiquitin binding / regulation of cytokinesis / N-glycan trimming in the ER and Calnexin/Calreticulin cycle / nucleotide-excision repair / DNA Damage Recognition in GG-NER / G-protein beta/gamma-subunit complex binding / microtubule cytoskeleton organization / Formation of Incision Complex in GG-NER / apical part of cell / Regulation of PLK1 Activity at G2/M Transition / protein transport / mitotic cell cycle / single-stranded DNA binding / microtubule binding / spermatogenesis / RNA polymerase II-specific DNA-binding transcription factor binding / proteasome-mediated ubiquitin-dependent protein catabolic process / damaged DNA binding / transcription coactivator activity / ciliary basal body / RNA polymerase II cis-regulatory region sequence-specific DNA binding / response to xenobiotic stimulus / cell division / DNA repair / intracellular membrane-bounded organelle / centrosome / calcium ion binding / protein-containing complex binding / chromatin / positive regulation of DNA-templated transcription / nucleolus / mitochondrion / nucleoplasm / nucleus / plasma membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 25.0 Å | |||||||||
 Authors Authors | Zhang ET / He Y / Grob P / Fong YW / Nogales E / Tjian R | |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2015 Journal: Proc Natl Acad Sci U S A / Year: 2015Title: Architecture of the human XPC DNA repair and stem cell coactivator complex. Authors: Elisa T Zhang / Yuan He / Patricia Grob / Yick W Fong / Eva Nogales / Robert Tjian /  Abstract: The Xeroderma pigmentosum complementation group C (XPC) complex is a versatile factor involved in both nucleotide excision repair and transcriptional coactivation as a critical component of the ...The Xeroderma pigmentosum complementation group C (XPC) complex is a versatile factor involved in both nucleotide excision repair and transcriptional coactivation as a critical component of the NANOG, OCT4, and SOX2 pluripotency gene regulatory network. Here we present the structure of the human holo-XPC complex determined by single-particle electron microscopy to reveal a flexible, ear-shaped structure that undergoes localized loss of order upon DNA binding. We also determined the structure of the complete yeast homolog Rad4 holo-complex to find a similar overall architecture to the human complex, consistent with their shared DNA repair functions. Localized differences between these structures reflect an intriguing phylogenetic divergence in transcriptional capabilities that we present here. Having positioned the constituent subunits by tagging and deletion, we propose a model of key interaction interfaces that reveals the structural basis for this difference in functional conservation. Together, our findings establish a framework for understanding the structure-function relationships of the XPC complex in the interplay between transcription and DNA repair. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6495.map.gz emd_6495.map.gz | 6.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6495-v30.xml emd-6495-v30.xml emd-6495.xml emd-6495.xml | 13.6 KB 13.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_6495.tif emd_6495.tif | 755.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6495 http://ftp.pdbj.org/pub/emdb/structures/EMD-6495 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6495 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6495 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_6495.map.gz / Format: CCP4 / Size: 7.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6495.map.gz / Format: CCP4 / Size: 7.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EMAN2 reconstruction of apo human XPC complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.01 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
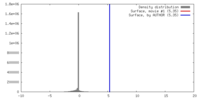
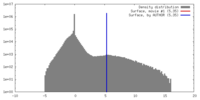
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Apo human XPC complex
| Entire | Name: Apo human XPC complex |
|---|---|
| Components |
|
-Supramolecule #1000: Apo human XPC complex
| Supramolecule | Name: Apo human XPC complex / type: sample / ID: 1000 Details: Monodisperse. Thawed from -80 degrees Celsius and placed on ice immediately prior to grid preparation. Oligomeric state: heterotrimer / Number unique components: 3 |
|---|---|
| Molecular weight | Experimental: 200 KDa / Theoretical: 200 KDa / Method: SDS-PAGE, size exclusion chromatography |
-Macromolecule #1: Xeroderma pigmentosum group C-complementing protein
| Macromolecule | Name: Xeroderma pigmentosum group C-complementing protein / type: protein_or_peptide / ID: 1 Name.synonym: XPC, p125, DNA repair protein complementing XP-C cells Details: contains N-terminal 6xHis and TEV cleavage site / Number of copies: 1 / Oligomeric state: monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: human / Location in cell: nucleus, cytoplasm Homo sapiens (human) / synonym: human / Location in cell: nucleus, cytoplasm |
| Molecular weight | Experimental: 125 KDa / Theoretical: 125 KDa |
| Recombinant expression | Organism:  |
| Sequence | UniProtKB: DNA repair protein complementing XP-C cells / GO: XPC complex |
-Macromolecule #2: RAD23 homolog B
| Macromolecule | Name: RAD23 homolog B / type: protein_or_peptide / ID: 2 Name.synonym: RAD23B, HR23B, HHR23B, p58, XP-C repair-complementing complex 58 kDa protein Details: contains N-terminal 1xFLAG tag / Number of copies: 1 / Oligomeric state: monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: human / Location in cell: nucleus, cytoplasm Homo sapiens (human) / synonym: human / Location in cell: nucleus, cytoplasm |
| Molecular weight | Experimental: 58 KDa / Theoretical: 58 KDa |
| Recombinant expression | Organism:  |
| Sequence | UniProtKB: UV excision repair protein RAD23 homolog B / GO: XPC complex / InterPro: UV excision repair protein Rad23 |
-Macromolecule #3: Centrin-2
| Macromolecule | Name: Centrin-2 / type: protein_or_peptide / ID: 3 / Name.synonym: CETN2 / Number of copies: 1 / Oligomeric state: monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: human / Location in cell: nucleus, cytoplasm, centrosomes Homo sapiens (human) / synonym: human / Location in cell: nucleus, cytoplasm, centrosomes |
| Molecular weight | Experimental: 18 KDa / Theoretical: 18 KDa |
| Recombinant expression | Organism:  |
| Sequence | UniProtKB: Centrin-2 / GO: XPC complex / InterPro: INTERPRO: IPR029528 |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.01 mg/mL |
|---|---|
| Buffer | pH: 7.6 Details: 300 mM KCl, 50 mM HEPES, 0.1% NP-40 alternative, 10% glycerol, 0.1 mM EDTA, 1 mM MgCl2, 1 mM TCEP, 1 mM DTT |
| Staining | Type: NEGATIVE Details: Grids with adsorbed protein were floated on 2 successive droplets of buffer G (300 mM KCl, 25 mM HEPES ph 7.6, 3% w/v trehalose, 0.01% NP-40 alternative, 1 mM TCEP, 1 mM DTT, 0.1 mM EDTA, 1 ...Details: Grids with adsorbed protein were floated on 2 successive droplets of buffer G (300 mM KCl, 25 mM HEPES ph 7.6, 3% w/v trehalose, 0.01% NP-40 alternative, 1 mM TCEP, 1 mM DTT, 0.1 mM EDTA, 1 mM MgCl2) and subsequently floated on 4 successive 1% w/v uranyl droplets for 10 seconds each. |
| Grid | Details: 400 mesh copper grid with thin carbon support |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Alignment procedure | Legacy - Astigmatism: Astigmatism was corrected at 80,000 times and 280,000 times magnification. |
| Date | Aug 13, 2013 |
| Image recording | Category: CCD / Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Number real images: 426 / Average electron dose: 20 e/Å2 / Bits/pixel: 32 |
| Tilt angle min | 0 |
| Tilt angle max | 0 |
| Electron beam | Acceleration voltage: 120 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.2 mm / Nominal defocus max: 2.25 µm / Nominal defocus min: -0.07 µm / Nominal magnification: 80000 |
| Sample stage | Specimen holder model: OTHER |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | Particles were selected by DoGPicker in the Appion pipeline. |
|---|---|
| CTF correction | Details: whole micrograph |
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 25.0 Å / Resolution method: OTHER / Software - Name: EMAN2 / Number images used: 22777 |
| Final two d classification | Number classes: 141 |
 Movie
Movie Controller
Controller





















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)