+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3307 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the human TAF-less PIC in the closed state | |||||||||
 Map data Map data | Reconstruction of the human TAF-less PIC | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | TFIID / TFIIA / transcription / RNA polymerase II / general transcription factors / preinitiation complex / core promoter / DNA binding | |||||||||
| Function / homology |  Function and homology information Function and homology informationmicrofibril binding / MMXD complex / core TFIIH complex portion of holo TFIIH complex / Cytosolic iron-sulfur cluster assembly / central nervous system myelin formation / RNA Polymerase III Chain Elongation / positive regulation of core promoter binding / RNA polymerase II core complex assembly / RNA polymerase II CTD heptapeptide repeat S5 kinase activity / positive regulation of mitotic recombination ...microfibril binding / MMXD complex / core TFIIH complex portion of holo TFIIH complex / Cytosolic iron-sulfur cluster assembly / central nervous system myelin formation / RNA Polymerase III Chain Elongation / positive regulation of core promoter binding / RNA polymerase II core complex assembly / RNA polymerase II CTD heptapeptide repeat S5 kinase activity / positive regulation of mitotic recombination / hair cell differentiation / hair follicle maturation / RNA Polymerase III Transcription Termination / transcription factor TFIIE complex / nucleotide-excision repair factor 3 complex / RNA polymerase transcription factor SL1 complex / nucleotide-excision repair, preincision complex assembly / meiotic sister chromatid cohesion / phosphatase activator activity / ventricular system development / snRNA transcription by RNA polymerase II / CAK-ERCC2 complex / RNA polymerase III general transcription initiation factor activity / TFIIF-class transcription factor complex binding / regulation of transcription by RNA polymerase I / transcription factor TFIIK complex / transcriptional start site selection at RNA polymerase II promoter / RNA polymerase I core promoter sequence-specific DNA binding / RPAP3/R2TP/prefoldin-like complex / transcription factor TFIIF complex / embryonic cleavage / DNA 5'-3' helicase / RNA Polymerase III Transcription Initiation From Type 1 Promoter / RNA Polymerase III Transcription Initiation From Type 2 Promoter / RNA Polymerase III Transcription Initiation From Type 3 Promoter / transcription factor TFIIA complex / female germ cell nucleus / UV protection / regulation of cyclin-dependent protein serine/threonine kinase activity / adult heart development / RNA Polymerase III Abortive And Retractive Initiation / Cytosolic sensors of pathogen-associated DNA / transcription factor TFIIH core complex / transcription factor TFIIH holo complex / male pronucleus / cyclin-dependent protein serine/threonine kinase activator activity / female pronucleus / G protein-coupled receptor internalization / germinal vesicle / RNA polymerase II general transcription initiation factor binding / nuclear thyroid hormone receptor binding / [RNA-polymerase]-subunit kinase / Abortive elongation of HIV-1 transcript in the absence of Tat / transcription preinitiation complex / FGFR2 alternative splicing / RNA Polymerase I Transcription Termination / cyclin-dependent protein serine/threonine kinase regulator activity / MicroRNA (miRNA) biogenesis / Viral Messenger RNA Synthesis / Signaling by FGFR2 IIIa TM / LRR domain binding / protein acetylation / RNA polymerase II general transcription initiation factor activity / transcription factor TFIID complex / regulation of mitotic cell cycle phase transition / cell division site / erythrocyte maturation / hematopoietic stem cell proliferation / acetyltransferase activity / spinal cord development / RNA Pol II CTD phosphorylation and interaction with CE during HIV infection / RNA Pol II CTD phosphorylation and interaction with CE / positive regulation of nuclear-transcribed mRNA poly(A) tail shortening / 3'-5' DNA helicase activity / HIV Transcription Initiation / RNA Polymerase II HIV Promoter Escape / Transcription of the HIV genome / RNA Polymerase II Promoter Escape / RNA Polymerase II Transcription Pre-Initiation And Promoter Opening / RNA Polymerase II Transcription Initiation / RNA Polymerase II Transcription Initiation And Promoter Clearance / Formation of the Early Elongation Complex / Formation of the HIV-1 Early Elongation Complex / bone mineralization / mRNA Capping / DNA 3'-5' helicase / mRNA Splicing - Minor Pathway / PIWI-interacting RNA (piRNA) biogenesis / aryl hydrocarbon receptor binding / histone acetyltransferase activity / viral transcription / TFIIB-class transcription factor binding / RNA polymerase II complex binding / ATPase activator activity / DNA topological change / intrinsic apoptotic signaling pathway by p53 class mediator / RNA Polymerase I Transcription Initiation / maintenance of transcriptional fidelity during transcription elongation by RNA polymerase II / Processing of Capped Intron-Containing Pre-mRNA / transcription by RNA polymerase III Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
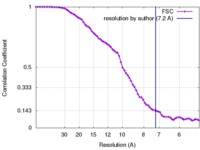
| Method | single particle reconstruction / cryo EM / Resolution: 7.2 Å | |||||||||
 Authors Authors | Louder RK / He Y / Lopez-Blanco JR / Fang J / Chacon P / Nogales E | |||||||||
 Citation Citation |  Journal: Nature / Year: 2016 Journal: Nature / Year: 2016Title: Structure of promoter-bound TFIID and model of human pre-initiation complex assembly. Authors: Robert K Louder / Yuan He / José Ramón López-Blanco / Jie Fang / Pablo Chacón / Eva Nogales /   Abstract: The general transcription factor IID (TFIID) plays a central role in the initiation of RNA polymerase II (Pol II)-dependent transcription by nucleating pre-initiation complex (PIC) assembly at the ...The general transcription factor IID (TFIID) plays a central role in the initiation of RNA polymerase II (Pol II)-dependent transcription by nucleating pre-initiation complex (PIC) assembly at the core promoter. TFIID comprises the TATA-binding protein (TBP) and 13 TBP-associated factors (TAF1-13), which specifically interact with a variety of core promoter DNA sequences. Here we present the structure of human TFIID in complex with TFIIA and core promoter DNA, determined by single-particle cryo-electron microscopy at sub-nanometre resolution. All core promoter elements are contacted by subunits of TFIID, with TAF1 and TAF2 mediating major interactions with the downstream promoter. TFIIA bridges the TBP-TATA complex with lobe B of TFIID. We also present the cryo-electron microscopy reconstruction of a fully assembled human TAF-less PIC. Superposition of common elements between the two structures provides novel insights into the general role of TFIID in promoter recognition, PIC assembly, and transcription initiation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3307.map.gz emd_3307.map.gz | 1.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3307-v30.xml emd-3307-v30.xml emd-3307.xml emd-3307.xml | 20 KB 20 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_3307_fsc.xml emd_3307_fsc.xml | 8 KB | Display |  FSC data file FSC data file |
| Images |  emd_3307.png emd_3307.png | 64.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3307 http://ftp.pdbj.org/pub/emdb/structures/EMD-3307 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3307 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3307 | HTTPS FTP |
-Related structure data
| Related structure data |  5iy6M  6o9lM  3304C  3305C  3306C  5furC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_3307.map.gz / Format: CCP4 / Size: 26.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3307.map.gz / Format: CCP4 / Size: 26.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of the human TAF-less PIC | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.64 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 Movie
Movie Controller
Controller Sample components
Sample components



 UCSF Chimera
UCSF Chimera














































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)






















 Processing
Processing Electron microscopy
Electron microscopy