+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-31912 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | 2.26 A structure of the glutamate dehydrogenase | ||||||||||||||||||
 Map data Map data | |||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | Glutamate dehydrogenase / STRUCTURAL PROTEIN | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationL-glutamate dehydrogenase [NAD(P)+] activity / tricarboxylic acid metabolic process / glutamate dehydrogenase [NAD(P)+] / L-glutamate dehydrogenase (NADP+) activity / L-glutamate dehydrogenase (NAD+) activity / L-glutamate catabolic process / glutamine metabolic process / mitochondrial inner membrane / GTP binding / endoplasmic reticulum ...L-glutamate dehydrogenase [NAD(P)+] activity / tricarboxylic acid metabolic process / glutamate dehydrogenase [NAD(P)+] / L-glutamate dehydrogenase (NADP+) activity / L-glutamate dehydrogenase (NAD+) activity / L-glutamate catabolic process / glutamine metabolic process / mitochondrial inner membrane / GTP binding / endoplasmic reticulum / mitochondrion / ATP binding / identical protein binding Similarity search - Function | ||||||||||||||||||
| Biological species |  | ||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.26 Å | ||||||||||||||||||
 Authors Authors | Fan HC / Zhang Y | ||||||||||||||||||
| Funding support |  China, 5 items China, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: A cryo-electron microscopy support film formed by 2D crystals of hydrophobin HFBI. Authors: Hongcheng Fan / Bo Wang / Yan Zhang / Yun Zhu / Bo Song / Haijin Xu / Yujia Zhai / Mingqiang Qiao / Fei Sun /  Abstract: Cryo-electron microscopy (cryo-EM) has become a powerful tool to resolve high-resolution structures of biomacromolecules in solution. However, air-water interface induced preferred orientations, ...Cryo-electron microscopy (cryo-EM) has become a powerful tool to resolve high-resolution structures of biomacromolecules in solution. However, air-water interface induced preferred orientations, dissociation or denaturation of biomacromolecules during cryo-vitrification remains a limiting factor for many specimens. To solve this bottleneck, we developed a cryo-EM support film using 2D crystals of hydrophobin HFBI. The hydrophilic side of the HFBI film adsorbs protein particles via electrostatic interactions and sequesters them from the air-water interface, allowing the formation of sufficiently thin ice for high-quality data collection. The particle orientation distribution can be regulated by adjusting the buffer pH. Using this support, we determined the cryo-EM structures of catalase (2.29 Å) and influenza haemagglutinin trimer (2.56 Å), which exhibited strong preferred orientations using a conventional cryo-vitrification protocol. We further show that the HFBI film is suitable to obtain high-resolution structures of small proteins, including aldolase (150 kDa, 3.28 Å) and haemoglobin (64 kDa, 3.6 Å). Our work suggests that HFBI films may have broad future applications in increasing the success rate and efficiency of cryo-EM. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_31912.map.gz emd_31912.map.gz | 228.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-31912-v30.xml emd-31912-v30.xml emd-31912.xml emd-31912.xml | 19 KB 19 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_31912_fsc.xml emd_31912_fsc.xml | 14.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_31912.png emd_31912.png | 157.1 KB | ||
| Filedesc metadata |  emd-31912.cif.gz emd-31912.cif.gz | 6.1 KB | ||
| Others |  emd_31912_half_map_1.map.gz emd_31912_half_map_1.map.gz emd_31912_half_map_2.map.gz emd_31912_half_map_2.map.gz | 194 MB 194.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-31912 http://ftp.pdbj.org/pub/emdb/structures/EMD-31912 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31912 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31912 | HTTPS FTP |
-Validation report
| Summary document |  emd_31912_validation.pdf.gz emd_31912_validation.pdf.gz | 1.2 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_31912_full_validation.pdf.gz emd_31912_full_validation.pdf.gz | 1.2 MB | Display | |
| Data in XML |  emd_31912_validation.xml.gz emd_31912_validation.xml.gz | 20.8 KB | Display | |
| Data in CIF |  emd_31912_validation.cif.gz emd_31912_validation.cif.gz | 27.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31912 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31912 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31912 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31912 | HTTPS FTP |
-Related structure data
| Related structure data |  7vdaMC  7vd8C  7vd9C  7vdcC  7vdeC  7vdfC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_31912.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_31912.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.65 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
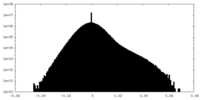
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: #2
| File | emd_31912_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
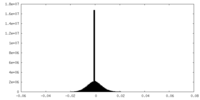
| Density Histograms |
-Half map: #1
| File | emd_31912_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
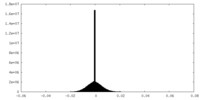
| Density Histograms |
- Sample components
Sample components
-Entire : Glutamate dehydrogenase
| Entire | Name: Glutamate dehydrogenase |
|---|---|
| Components |
|
-Supramolecule #1: Glutamate dehydrogenase
| Supramolecule | Name: Glutamate dehydrogenase / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 334 KDa |
-Macromolecule #1: Glutamate dehydrogenase 1, mitochondrial
| Macromolecule | Name: Glutamate dehydrogenase 1, mitochondrial / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO / EC number: glutamate dehydrogenase [NAD(P)+] |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 61.60891 KDa |
| Sequence | String: MYRYLGEALL LSRAGPAALG SASADSAALL GWARGQPAAA PQPGLVPPAR RHYSEAAADR EDDPNFFKMV EGFFDRGASI VEDKLVEDL KTRETEEQKR NRVRSILRII KPCNHVLSLS FPIRRDDGSW EVIEGYRAQH SQHRTPCKGG IRYSTDVSVD E VKALASLM ...String: MYRYLGEALL LSRAGPAALG SASADSAALL GWARGQPAAA PQPGLVPPAR RHYSEAAADR EDDPNFFKMV EGFFDRGASI VEDKLVEDL KTRETEEQKR NRVRSILRII KPCNHVLSLS FPIRRDDGSW EVIEGYRAQH SQHRTPCKGG IRYSTDVSVD E VKALASLM TYKCAVVDVP FGGAKAGVKI NPKNYTDNEL EKITRRFTME LAKKGFIGPG VDVPAPDMST GEREMSWIAD TY ASTIGHY DINAHACVTG KPISQGGIHG RISATGRGVF HGIENFINEA SYMSILGMTP GFGDKTFVVQ GFGNVGLHSM RYL HRFGAK CITVGESDGS IWNPDGIDPK ELEDFKLQHG TILGFPKAKI YEGSILEVDC DILIPAASEK QLTKSNAPRV KAKI IAEGA NGPTTPEADK IFLERNIMVI PDLYLNAGGV TVSYFEWLKN LNHVSYGRLT FKYERDSNYH LLMSVQESLE RKFGK HGGT IPIVPTAEFQ DRISGASEKD IVHSGLAYTM ERSARQIMRT AMKYNLGLDL RTAAYVNAIE KVFRVYNEAG VTFT UniProtKB: Glutamate dehydrogenase 1, mitochondrial |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Component - Formula: PBS |
| Grid | Material: NICKEL/TITANIUM / Mesh: 300 |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: SUPER-RESOLUTION / Number real images: 1635 / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal magnification: 215000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)