+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 5tao | ||||||
|---|---|---|---|---|---|---|---|
| Title | Haloferax volcanii Malate Synthase Lead(II) complex | ||||||
 Components Components | Malate synthase | ||||||
 Keywords Keywords | TRANSFERASE | ||||||
| Function / homology |  Function and homology information Function and homology informationmalate synthase / malate synthase activity / oxaloacetate metabolic process / glyoxylate cycle / tricarboxylic acid cycle / magnesium ion binding / metal ion binding / cytoplasm Similarity search - Function | ||||||
| Biological species |  Haloferax volcanii (archaea) Haloferax volcanii (archaea) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / MOLECULAR REPLACEMENT /  molecular replacement / Resolution: 2.1 Å molecular replacement / Resolution: 2.1 Å | ||||||
 Authors Authors | Howard, B.R. / Adams, M.J. | ||||||
 Citation Citation |  Journal: The Journal of the Utah Academy / Year: 2016 Journal: The Journal of the Utah Academy / Year: 2016Title: X-ray analysis of Lead(II) binding to Haloferax volcanii Malate Synthase Authors: Adams, M.J. / Howard, B.R. #1:  Journal: BMC Struct. Biol. / Year: 2011 Journal: BMC Struct. Biol. / Year: 2011Title: Crystal structures of a halophilic archaeal malate synthase from Haloferax volcanii and comparisons with isoforms A and G. Authors: Bracken, C.D. / Neighbor, A.M. / Lamlenn, K.K. / Thomas, G.C. / Schubert, H.L. / Whitby, F.G. / Howard, B.R. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  5tao.cif.gz 5tao.cif.gz | 98.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb5tao.ent.gz pdb5tao.ent.gz | 72 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  5tao.json.gz 5tao.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ta/5tao https://data.pdbj.org/pub/pdb/validation_reports/ta/5tao ftp://data.pdbj.org/pub/pdb/validation_reports/ta/5tao ftp://data.pdbj.org/pub/pdb/validation_reports/ta/5tao | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  3pugS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | x 6
| ||||||||||||
| 2 | 
| ||||||||||||
| Unit cell |
| ||||||||||||
| Components on special symmetry positions |
|
- Components
Components
-Protein , 1 types, 1 molecules A
| #1: Protein | Mass: 48066.848 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Haloferax volcanii (strain ATCC 29605 / DSM 3757 / JCM 8879 / NBRC 14742 / NCIMB 2012 / VKM B-1768 / DS2) (archaea) Haloferax volcanii (strain ATCC 29605 / DSM 3757 / JCM 8879 / NBRC 14742 / NCIMB 2012 / VKM B-1768 / DS2) (archaea)Strain: ATCC 29605 / DSM 3757 / JCM 8879 / NBRC 14742 / NCIMB 2012 / VKM B-1768 / DS2 Gene: aceB, aceB1, HVO_1983, C498_05196 / Production host:  Haloferax volcanii (archaea) / References: UniProt: D4GTL2, malate synthase Haloferax volcanii (archaea) / References: UniProt: D4GTL2, malate synthase |
|---|
-Non-polymers , 7 types, 270 molecules 
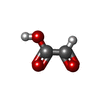











| #2: Chemical | ChemComp-ACT / | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| #3: Chemical | | #4: Chemical | ChemComp-PB / #5: Chemical | #6: Chemical | ChemComp-CL / #7: Chemical | ChemComp-NA / | #8: Water | ChemComp-HOH / | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.37 Å3/Da / Density % sol: 63.48 % |
|---|---|
| Crystal grow | Temperature: 298 K / Method: vapor diffusion, sitting drop Details: magnesium chloride, glyoxylate, Tris, KCl, ammonium acetate, PEG 4500, glycerol, sodium acetate trihydrate PH range: 4.4-5.0 |
-Data collection
| Diffraction | Mean temperature: 100 K | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU MICROMAX-007 HF / Wavelength: 1.54 Å ROTATING ANODE / Type: RIGAKU MICROMAX-007 HF / Wavelength: 1.54 Å | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Detector | Type: RIGAKU RAXIS IV / Detector: IMAGE PLATE / Date: Mar 15, 2009 / Details: Rigaku Varimax-HR | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation wavelength | Wavelength: 1.54 Å / Relative weight: 1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection | Resolution: 2.1→96.81 Å / Num. obs: 35791 / % possible obs: 99.5 % / Redundancy: 3.7 % / Rmerge(I) obs: 0.103 / Net I/av σ(I): 11.453 / Net I/σ(I): 13.2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection shell |
|
-Phasing
| Phasing | Method:  molecular replacement molecular replacement | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Phasing MR | Model details: Phaser MODE: MR_AUTO
|
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 3PUG Resolution: 2.1→96.81 Å / Cor.coef. Fo:Fc: 0.954 / Cor.coef. Fo:Fc free: 0.941 / SU B: 3.507 / SU ML: 0.094 / Cross valid method: THROUGHOUT / ESU R: 0.143 / ESU R Free: 0.137 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å / Solvent model: BABINET MODEL WITH MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 48.764 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: 1 / Resolution: 2.1→96.81 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller










 PDBj
PDBj









