+ Open data
Open data
 Loading...
Loading...
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23937 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the human SSU processome, state pre-A1* | |||||||||||||||
 Map data Map data | Local resolution filtered map | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Ribosomal assembly intermediate / RIBOSOME | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationmRNA N-acetyltransferase activity / U6 snRNA 2'-O-ribose methyltransferase activity / oocyte growth / nucleologenesis / snoRNA localization / granular component / leucine zipper domain binding / tRNA wobble cytosine modification / U4atac snRNP / tRNA cytidine N4-acetyltransferase activity ...mRNA N-acetyltransferase activity / U6 snRNA 2'-O-ribose methyltransferase activity / oocyte growth / nucleologenesis / snoRNA localization / granular component / leucine zipper domain binding / tRNA wobble cytosine modification / U4atac snRNP / tRNA cytidine N4-acetyltransferase activity / rRNA acetylation involved in maturation of SSU-rRNA / 18S rRNA cytidine N-acetyltransferase activity / regulation of stem cell population maintenance / tRNA acetylation / U4atac snRNA binding / CURI complex / UTP-C complex / negative regulation of amyloid precursor protein biosynthetic process / pre-snoRNP complex / t-UTP complex / Pwp2p-containing subcomplex of 90S preribosome / Mpp10 complex / rRNA (pseudouridine) methyltransferase activity / box C/D sno(s)RNA binding / rRNA modification / histone H2AQ104 methyltransferase activity / preribosome / dense fibrillar component / box C/D sno(s)RNA 3'-end processing / histone methyltransferase binding / rRNA methyltransferase activity / endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / regulation of centrosome duplication / regulation of transcription elongation by RNA polymerase II / endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / N-acetyltransferase activity / cilium disassembly / box C/D methylation guide snoRNP complex / embryonic cleavage / positive regulation of rRNA processing / tRNA export from nucleus / RNA splicing, via transesterification reactions / rRNA primary transcript binding / transcription elongation factor activity / sno(s)RNA-containing ribonucleoprotein complex / blastocyst formation / rRNA base methylation / SUMOylation of RNA binding proteins / U4 snRNA binding / U2-type precatalytic spliceosome / protein localization to nucleolus / telomerase holoenzyme complex / spindle assembly involved in female meiosis / rRNA methylation / box C/D snoRNP assembly / epigenetic programming in the zygotic pronuclei / neural precursor cell proliferation / negative regulation of RNA splicing / U3 snoRNA binding / neural crest cell differentiation / protein acetylation / NRAGE signals death through JNK / negative regulation of bicellular tight junction assembly / snoRNA binding / precatalytic spliceosome / preribosome, small subunit precursor / rRNA modification in the nucleus and cytosol / erythrocyte homeostasis / Formation of the ternary complex, and subsequently, the 43S complex / cytoplasmic side of rough endoplasmic reticulum membrane / rRNA metabolic process / negative regulation of ubiquitin protein ligase activity / Cul4-RING E3 ubiquitin ligase complex / Ribosomal scanning and start codon recognition / Association of TriC/CCT with target proteins during biosynthesis / positive regulation of transcription by RNA polymerase I / Translation initiation complex formation / negative regulation of telomere maintenance via telomerase / decidualization / TFIID-class transcription factor complex binding / RNA polymerase II complex binding / TOR signaling / Protein hydroxylation / SARS-CoV-1 modulates host translation machinery / mTORC1-mediated signalling / negative regulation of apoptotic signaling pathway / Peptide chain elongation / cellular response to ethanol / Selenocysteine synthesis / Formation of a pool of free 40S subunits / positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator / Eukaryotic Translation Termination / blastocyst development / SRP-dependent cotranslational protein targeting to membrane / Response of EIF2AK4 (GCN2) to amino acid deficiency / negative regulation of ubiquitin-dependent protein catabolic process / ubiquitin ligase inhibitor activity / Viral mRNA Translation / 90S preribosome Similarity search - Function Protein of unknown function DUF4602 / : / Nucleolar protein 7, C-terminal / Rrp7A, RNA recognition motif / : / NUC129 domain / U3 small nucleolar RNA-associated protein 6 C-terminal / 40S small subunit processome assembly factor 1 / WD repeat-containing protein 75 first beta-propeller / Apoptosis-antagonizing transcription factor, C-terminal ...Protein of unknown function DUF4602 / : / Nucleolar protein 7, C-terminal / Rrp7A, RNA recognition motif / : / NUC129 domain / U3 small nucleolar RNA-associated protein 6 C-terminal / 40S small subunit processome assembly factor 1 / WD repeat-containing protein 75 first beta-propeller / Apoptosis-antagonizing transcription factor, C-terminal / AATF leucine zipper-containing domain / Protein AATF/Bfr2 / Apoptosis-antagonizing transcription factor, C-terminal / Apoptosis antagonizing transcription factor / NUC153 / Nucleolar protein 10/Enp2 / : / : / NUC153 domain / Nucleolar protein 10-like, second domain / Nucleolar protein 10-like, N-terminal domain / Ribosomal RNA assembly KRR1 / : / : / : / KRR1 small subunit processome component, second KH domain / Possible tRNA binding domain / RNA cytidine acetyltransferase NAT10 / Possible tRNA binding domain / Helicase domain / tRNA(Met) cytidine acetyltransferase TmcA, N-terminal / TmcA/NAT10/Kre33 / RNA cytidine acetyltransferase NAT10/TcmA, helicase domain / tRNA(Met) cytidine acetyltransferase TmcA, N-terminal / GNAT acetyltransferase 2 / Sof1-like protein / U3 small nucleolar RNA-associated protein 20, N-terminal / U3 small nucleolar RNA-associated protein 20, C-terminal / : / : / Sof1-like domain / U3 small nucleolar RNA-associated protein 20, N-terminal / U3 small nucleolar RNA-associated protein 20 domain / Small subunit processome component 20 homolog, C-terminal / WD repeat-containing protein 75 second beta-propeller / NOL6/Upt22 / BING4, C-terminal domain / Nrap protein domain 1 / Nrap protein, domain 2 / Nrap protein, domain 3 / Nrap protein, domain 4 / Nrap protein, domain 5 / Nrap protein, domain 6 / WD repeat-containing protein WDR46/Utp7 / Nrap protein domain 1 / BING4CT (NUC141) domain / Nrap protein PAP/OAS-like domain / Nrap protein domain 3 / Nrap protein nucleotidyltransferase domain 4 / Nrap protein PAP/OAS1-like domain 5 / Nrap protein domain 6 / BING4CT (NUC141) domain / U3 small nucleolar RNA-associated protein 6 / Ribosomal RNA-processing protein 7, C-terminal domain / Ribosomal RNA-processing protein 7 / Rrp7, RRM-like N-terminal domain / : / U3 small nucleolar RNA-associated protein 6 / Ribosomal RNA-processing protein 7 (RRP7) C-terminal domain / Rrp7 RRM-like N-terminal domain / U3 small nucleolar RNA-associated protein 4 / Small-subunit processome, Utp11 / Fcf2 pre-rRNA processing, C-terminal / Fcf2/DNTTIP2 / : / Utp11 protein / Fcf2 pre-rRNA processing / : / U3 small nucleolar RNA-associated protein 15, C-terminal / UTP15 C terminal / rRNA-processing protein Fcf1, PIN domain / WDR3 second beta-propeller domain / WDR3 first beta-propeller domain / Small-subunit processome, Utp14 / U3 small nucleolar RNA-associated protein 18 / Utp14 protein / U3 small nucleolar RNA-associated protein 13, C-terminal / Periodic tryptophan protein 2 / Utp13 specific WD40 associated domain / BP28, C-terminal domain / Nucleolar protein 58/56, N-terminal / U3 small nucleolar RNA-associated protein 10, N-terminal / U3 small nucleolar RNA-associated protein 10 / : / BP28CT (NUC211) domain / NOP5NT (NUC127) domain / U3 small nucleolar RNA-associated protein 10 / HEATR1-like, HEAT repeats / BP28CT (NUC211) domain / rRNA-processing protein Fcf1/Utp23 Similarity search - Domain/homology U3 small nucleolar ribonucleoprotein protein MPP10 / Nucleolar protein 56 / WD repeat-containing protein 46 / U3 small nucleolar RNA-interacting protein 2 / Small subunit processome component 20 homolog / Small ribosomal subunit protein eS17 / rRNA 2'-O-methyltransferase fibrillarin / Small ribosomal subunit protein eS12 / Small ribosomal subunit protein eS27 / Small ribosomal subunit protein uS4 ...U3 small nucleolar ribonucleoprotein protein MPP10 / Nucleolar protein 56 / WD repeat-containing protein 46 / U3 small nucleolar RNA-interacting protein 2 / Small subunit processome component 20 homolog / Small ribosomal subunit protein eS17 / rRNA 2'-O-methyltransferase fibrillarin / Small ribosomal subunit protein eS12 / Small ribosomal subunit protein eS27 / Small ribosomal subunit protein uS4 / Small ribosomal subunit protein uS7 / NHP2-like protein 1 / Small ribosomal subunit protein eS1 / Small ribosomal subunit protein eS7 / Small ribosomal subunit protein eS8 / Small ribosomal subunit protein uS8 / Small ribosomal subunit protein uS9 / Small ribosomal subunit protein uS11 / Small ribosomal subunit protein uS12 / Small ribosomal subunit protein uS15 / Small ribosomal subunit protein uS17 / Small ribosomal subunit protein eS6 / Small ribosomal subunit protein eS24 / Small ribosomal subunit protein eS28 / Ubiquitin-ribosomal protein eS31 fusion protein / Transducin beta-like protein 3 / KRR1 small subunit processome component homolog / Ribosome biogenesis protein BMS1 homolog / WD repeat-containing protein 43 / Periodic tryptophan protein 2 homolog / Deoxynucleotidyltransferase terminal-interacting protein 2 / WD repeat-containing protein 75 / 40S small subunit processome assembly factor 1 / WD repeat-containing protein 36 / U3 small nucleolar RNA-associated protein 15 homolog / Ribosomal RNA small subunit methyltransferase NEP1 / U3 small nucleolar RNA-associated protein 4 homolog / U3 small nucleolar ribonucleoprotein protein IMP4 / Nucleolar protein 10 / U3 small nucleolar RNA-associated protein 14 homolog A / RNA cytidine acetyltransferase / HEAT repeat-containing protein 1 / Nucleolar protein 6 / Something about silencing protein 10 / RNA-binding protein PNO1 / DDB1- and CUL4-associated factor 13 / U3 small nucleolar ribonucleoprotein protein IMP3 / Protein AATF / U3 small nucleolar RNA-associated protein 6 homolog / U3 small nucleolar RNA-associated protein NOL7 / WD repeat-containing protein 3 / RNA 3'-terminal phosphate cyclase-like protein / Nucleolar protein 58 / rRNA-processing protein FCF1 homolog / Probable U3 small nucleolar RNA-associated protein 11 / Ribosomal RNA-processing protein 7 homolog A / U3 small nucleolar RNA-associated protein 18 homolog Similarity search - Component | |||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.87 Å | |||||||||||||||
 Authors Authors | Vanden Broeck A / Singh S / Klinge S | |||||||||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||||||||
 Citation Citation |  Journal: Science / Year: 2021 Journal: Science / Year: 2021Title: Nucleolar maturation of the human small subunit processome. Authors: Sameer Singh / Arnaud Vanden Broeck / Linamarie Miller / Malik Chaker-Margot / Sebastian Klinge /  Abstract: The human small subunit processome mediates early maturation of the small ribosomal subunit by coupling RNA folding to subsequent RNA cleavage and processing steps. We report the high-resolution ...The human small subunit processome mediates early maturation of the small ribosomal subunit by coupling RNA folding to subsequent RNA cleavage and processing steps. We report the high-resolution cryo–electron microscopy structures of maturing human small subunit (SSU) processomes at resolutions of 2.7 to 3.9 angstroms. These structures reveal the molecular mechanisms that enable crucial progressions during SSU processome maturation. RNA folding states within these particles are communicated to and coordinated with key enzymes that drive irreversible steps such as targeted exosome-mediated RNA degradation, protein-guided site-specific endonucleolytic RNA cleavage, and tightly controlled RNA unwinding. These conserved mechanisms highlight the SSU processome’s impressive structural plasticity, which endows this 4.5-megadalton nucleolar assembly with the distinctive ability to mature the small ribosomal subunit from within. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23937.map.gz emd_23937.map.gz | 51 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23937-v30.xml emd-23937-v30.xml emd-23937.xml emd-23937.xml | 123.6 KB 123.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_23937_fsc.xml emd_23937_fsc.xml | 19.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_23937.png emd_23937.png | 160 KB | ||
| Masks |  emd_23937_msk_1.map emd_23937_msk_1.map | 669.9 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-23937.cif.gz emd-23937.cif.gz | 31.1 KB | ||
| Others |  emd_23937_additional_1.map.gz emd_23937_additional_1.map.gz emd_23937_half_map_1.map.gz emd_23937_half_map_1.map.gz emd_23937_half_map_2.map.gz emd_23937_half_map_2.map.gz | 627.8 MB 540.8 MB 540.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23937 http://ftp.pdbj.org/pub/emdb/structures/EMD-23937 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23937 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23937 | HTTPS FTP |
-Related structure data
| Related structure data |  7mq9MC  7mq8C  7mqaC  7mqjC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23937.map.gz / Format: CCP4 / Size: 669.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23937.map.gz / Format: CCP4 / Size: 669.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Local resolution filtered map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_23937_msk_1.map emd_23937_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
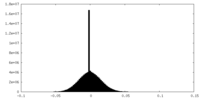
| Density Histograms |
-Additional map: Unmasked postprocessed map
| File | emd_23937_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unmasked postprocessed map | ||||||||||||
| Projections & Slices |
| ||||||||||||
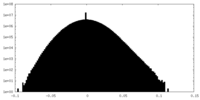
| Density Histograms |
-Half map: Half map 1
| File | emd_23937_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 2
| File | emd_23937_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : Human SSU processome
| Entire | Name: Human SSU processome |
|---|---|
| Components |
|
+Supramolecule #1: Human SSU processome
| Supramolecule | Name: Human SSU processome / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#69 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 5 MDa |
+Macromolecule #1: 5'ETS rRNA
| Macromolecule | Name: 5'ETS rRNA / type: rna / ID: 1 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 1.1668818749999998 MDa |
| Sequence | String: GCUGACACGC UGUCCUCUGG CGACCUGUCG CUGGAGAGGU UGGGCCUCCG GAUGCGCGCG GGGCUCUGGC CUACCGGUGA CCCGGCUAG CCGGCCGCGC UCCUGCUUGA GCCGCCUGCC GGGGCCCGCG GGCCUGCUGU UCUCUCGCGC GUCCGAGCGU C CCGACUCC ...String: GCUGACACGC UGUCCUCUGG CGACCUGUCG CUGGAGAGGU UGGGCCUCCG GAUGCGCGCG GGGCUCUGGC CUACCGGUGA CCCGGCUAG CCGGCCGCGC UCCUGCUUGA GCCGCCUGCC GGGGCCCGCG GGCCUGCUGU UCUCUCGCGC GUCCGAGCGU C CCGACUCC CGGUGCCGGC CCGGGUCCGG GUCUCUGACC CACCCGGGGG CGGCGGGGAA GGCGGCGAGG GCCACCGUGC CC CCGUGCG CUCUCCGCUG CGGGCGCCCG GGGCGGCCGC GACAACCCCA CCCCGCUGGC UCCGUGCCGU GCGUGUCAGG CGU UCUCGU CUCCGCGGGG UUGUCCGCCG CCCCUUCCCC GGAGUGGGGG GUUGGCCGGA GCCGAUCGGC UCGCUGGCCG GCCG GCCGG CCUCCGCUCC CGGGGGGCUC UUCGUGAUCG AUGUGGUGAC GUCGUGCUCU CCCGGGCCGG GUCCGAGCCG CGACG GGCG AGGGGCGGAC GUUCGUGGCG AACGGGACCG UCCUUCUCGC UCCGCCCCGC GGGGGUCCCC UCGUCUCUCC UCUCCC CGC CCGCCGGCGG UGCGUGUGGG AAGGCGUGGG GUGCGGACCC CGGCCCGACC UCGCCGUCCC GCCCGCCGCC UUCUGCG UC GCGGGUGCGG GCCGGCGGGG UCCUCUGACG CGGCAGACAG CCCUCGCUGU CGCCUCCAGU GGUUGUCGAC UUGCGGGC G GCCCCCCUCC GCGGCGGUGG GGGUGCCGUC CCGCCGGCCC GUCGUGCUGC CGUCUCGGGG GGUUUGCGCG AGCGUCGGC UCCGCCUGGG CCCUUGCGGU GCUCCUGGAG CGCUCCGGGU UGUCCCUCAG GUGCCCGAGG CCGAACGGUG GUGUGUCGUU CCCGCCCCC GGCGCCCCCU CCUCCGGUCG CCGCCGCGGU GUCCGCGCGU GGGUCCUGAG GGAGCUCGUC GGUGUGGGGU U CGAGGCGG UUUGAGUGAG ACGAGACGAG ACGCGCCCCU CCCACGCGGG GAAGGGCGCC CGCCUGCUCU CGGUGAGCGC AC GUCCCGU GCUCCCCUCU GGCGGGUGCG CGCGGGCCGU GUGAGCGAUC GCGGUGGGUU CGGGCCGGUG UGACGCGUGC GCC GGCCGG CCGCCGAGGG GCUGCCGUUC UGCCUCCGAC CGGUCGUGUG UGGGUUGACU UCGGAGGCGC UCUGCCUCGG AAGG AAGGA GGUGGGUGGA CGGGGGGGCC UGGUGGGGUU GCGCGCACGC GCGCACCGGC CGGGCCCCCG CCCUGAACGC GAACG CUCG AGGUGGCCGC GCGCAGGUGU UUCCUCGUAC CGCAGGGCCC CCUCCCUUCC CCAGGCGUCC CUCGGCGCCU CUGCGG GCC CGAGGAGGAG CGGCUGGCGG GUGGGGGGAG UGUGACCCAC CCUCGGUGAG AAAAGCCUUC UCUAGCGAUC UGAGAGG CG UGCCUUGGGG GUACCGGAUC CCCCGGGCCG CCGCCUCUGU CUCUGCCUCC GUUAUGGUAG CGCUGCCGUA GCGACCCG C UCGCAGAGGA CCCUCCUCCG CUUCCCCCUC GACGGGGUUG GGGGGGAGAA GCGAGGGUUC CGCCGGCCAC CGCGGUGGU GGCCGAGUGC GGCUCGUCGC CUACUGUGGC CCGCGCCUCC CCCUUCCGAG UCGGGGGAGG AUCCCGCCGG GCCGGGCCCG GCGUUCCCA GCGGGUUGGG ACGCGGCGGC CGGCGGGCGG UGGGUGUGCG CGCCCGGCGC UCUGUCCGGC GCGUGACCCC C UCCGCCGC GAGUCGGCUC UCCGCCCGCU CCCGUGCCGA GUCGUGACCG GUGCCGACGA CCGCGUUUGC GUGGCACGGG GU CGGGCCC GCCUGGCCCU GGGAAAGCGU CCCACGGUGG GGGCGCGCCG GUCUCCCGGA GCGGGACCGG GUCGGAGGAU GGA CGAGAA UCACGAGCGA CGGUGGUGGU GGCGUGUCGG GUUCGUGGCU GCGGUCGCUC CGGGGCCCCC GGUGGCGGGG CCCC GGGGC UCGCGAGGCG GUUCUCGGUG GGGGCCGAGG GCCGUCCGGC GUCCCAGGCG GGGCGCCGCG GGACCGCCCU CGUGU CUGU GGCGGUGGGA UCCCGCGGCC GUGUUUUCCU GGUGGCCCGG CCGUGCCUGA GGUUUCUCCC CGAGCCGCCG CCUCUG CGG GCUCCCGGGU GCCCUUGCCC UCGCGGUCCC CGGCCCUCGC CCGUCUGUGC CCUCUUCCCC GCCCGCCGCC CGCCGAU CC UCUUCUUCCC CCGAGCGGCU CACCGGCUUC ACGUCCGUUG GUGGCCCCGC CUGGGACCGA ACCCGGCACC GCCUCGUG G GGCGCCGCCG CCGGCCACUG AUCGGCCCGG CGUCCGCGUC CCCCGGCGCG CGCCUUGGGG ACCGGGUCGG UGGCGCCCC GCGUGGGGCC CGGUGGGCUU CCAGGAGGGU UCCGGGGGUC GGCCUGCGGC GCGUGCGGGG GAGGAGACGG UUCCGGGGGA CCGGCCGCG ACUGCGGCGG CGGUGGUGGG GGGAGCCGCG GGGAUCGCCG AGGGCCGGUC GGCCGCCCCG GGUGCCGCGC G GUGCCGCC GGCGGCGGUG AGGCCCCGCG CGUGUGUCCC GGCCGCGGUC GGCCGCGCUC GAGGGGUCCC CGUGGCGUCC CC UUCCCCG CCGGCCGCCU UUCUCGCGCC UUCCCCGUCG CCCCGGCCUC GCCCGUGGUC UCUCGUCUUC UCCCGGCCCG CUC UUCCGA ACCGGGUCGG CGCGUCCCCC GGGUGCGCCU CGCUUCCCGG GCCUGCCGCG GCCCUUCCCC GAGGCGUCCG UCCC GGGCG UCGGCGUCGG GGAGAGCCCG UCCUCCCCGC GUGGCGUCGC CCCGUUCGGC GCGCGCGUGC GCCCGAGCGC GGCCC GGUG GUCCCUCCCG GACAGGCGUU CGUGCGACGU GUGGCGUGGG UCGACCUCCG CCUUGCCGGU CGCUCGCCCU CUCCCC GGG UCGGGGGGUG GGGCCCGGGC CGGGGCCUCG GCCCCGGUCG CGGUCCCCCG UCCCGGGCGG GGGCGGGCGC GCCGGCC GG CCUCGGUCGC CCUCCCUUGG CCGUCGUGUG GCGUGUGCCA CCCCUGCGCC CGCGCCCGCC GGCGGGGCUC GGAGCCGG G CUUCGGCCGG GCCCCGGGCC CUCGACCGGA CCGGUGCGCG GGCGCUGCGG CCGCACGGCG CGACUGUCCC CGGGCCGGG CACCGCGGUC CGCCUCUCGC UCGCCGCCCG GACGUCGGGG CCGCCCCGCG GGGCGGGCGG AGCGCCGUCC CCGCCUCGCC GCCGCCCGC GGGCGCCGGC CGCGCGCGCG CGCGCGUGGC CGCCGGUCCC UCCCGGCCGC CGGGCGCGGG UCGGGCCGUC C GCCUCCUC GCGGGCGGGC GCGACGAAGA AGCGUCGCGG GUCUGUGGCG CGGGGCCCCC GGUGGUCGUG UCGCGUGGGG GG CGGGUGG UUGGGGCGUC CGGUUCGCCG CGCCCCGCCC CGGCCCCACC GGUCCCGGCC GCCGCCCCCG CGCCCGCUCG CUC CCUCCC GUCCGCCCGU CCGCGGCCCG UCCGUCCGUC CGUCCGUCGU CCUCCUCGCU UGCGGGGCGC CGGGCCCGUC CUCG ACGCG CUCUACCUUA CC GENBANK: GENBANK: MF164264.1 |
+Macromolecule #2: 18S rRNA
| Macromolecule | Name: 18S rRNA / type: rna / ID: 2 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 603.7575 KDa |
| Sequence | String: UACCUGGUUG AUCCUGCCAG UAGCAUAUGC UUGUCUCAAA GAUUAAGCCA UGCAUGUCUA AGUACGCACG GCCGGUACAG UGAAACUGC GAAUGGCUCA UUAAAUCAGU UAUGGUUCCU UUGGUCGCUC GCUCCUCUCC UACUUGGAUA ACUGUGGUAA U UCUAGAGC ...String: UACCUGGUUG AUCCUGCCAG UAGCAUAUGC UUGUCUCAAA GAUUAAGCCA UGCAUGUCUA AGUACGCACG GCCGGUACAG UGAAACUGC GAAUGGCUCA UUAAAUCAGU UAUGGUUCCU UUGGUCGCUC GCUCCUCUCC UACUUGGAUA ACUGUGGUAA U UCUAGAGC UAAUACAUGC CGACGGGCGC UGACCCCCUU CGCGGGGGGG AUGCGUGCAU UUAUCAGAUC AAAACCAACC CG GUCAGCC CCUCUCCGGC CCCGGCCGGG GGGCGGGCGC CGGCGGCUUU GGUGACUCUA GAUAACCUCG GGCCGAUCGC ACG CCCCCC GUGGCGGCGA CGACCCAUUC GAACGUCUGC CCUAUCAACU UUCGAUGGUA GUCGCCGUGC CUACCAUGGU GACC ACGGG UGACGGGGAA UCAGGGUUCG AUUCCGGAGA GGGAGCCUGA GAAACGGCUA CCACAUCCAA GGAAGGCAGC AGGCG CGCA AAUUACCCAC UCCCGACCCG GGGAGGUAGU GACGAAAAAU AACAAUACAG GACUCUUUCG AGGCCCUGUA AUUGGA AUG AGUCCACUUU AAAUCCUUUA ACGAGGAUCC AUUGGAGGGC AAGUCUGGUG CCAGCAGCCG CGGUAAUUCC AGCUCCA AU AGCGUAUAUU AAAGUUGCUG CAGUUAAAAA GCUCGUAGUU GGAUCUUGGG AGCGGGCGGG CGGUCCGCCG CGAGGCGA G CCACCGCCCG UCCCCGCCCC UUGCCUCUCG GCGCCCCCUC GAUGCUCUUA GCUGAGUGUC CCGCGGGGCC CGAAGCGUU UACUUUGAAA AAAUUAGAGU GUUCAAAGCA GGCCCGAGCC GCCUGGAUAC CGCAGCUAGG AAUAAUGGAA UAGGACCGCG GUUCUAUUU UGUUGGUUUU CGGAACUGAG GCCAUGAUUA AGAGGGACGG CCGGGGGCAU UCGUAUUGCG CCGCUAGAGG U GAAAUUCU UGGACCGGCG CAAGACGGAC CAGAGCGAAA GCAUUUGCCA AGAAUGUUUU CAUUAAUCAA GAACGAAAGU CG GAGGUUC GAAGACGAUC AGAUACCGUC GUAGUUCCGA CCAUAAACGA UGCCGACCGG CGAUGCGGCG GCGUUAUUCC CAU GACCCG CCGGGCAGCU UCCGGGAAAC CAAAGUCUUU GGGUUCCGGG GGGAGUAUGG UUGCAAAGCU GAAACUUAAA GGAA UUGAC GGAAGGGCAC CACCAGGAGU GGAGCCUGCG GCUUAAUUUG ACUCAACACG GGAAACCUCA CCCGGCCCGG ACACG GACA GGAUUGACAG AUUGAUAGCU CUUUCUCGAU UCCGUGGGUG GUGGUGCAUG GCCGUUCUUA GUUGGUGGAG CGAUUU GUC UGGUUAAUUC CGAUAACGAA CGAGACUCUG GCAUGCUAAC UAGUUACGCG ACCCCCGAGC GGUCGGCGUC CCCCAAC UU CUUAGAGGGA CAAGUGGCGU UCAGCCACCC GAGAUUGAGC AAUAACAGGU CUGUGAUGCC CUUAGAUGUC CGGGGCUG C ACGCGCGCUA CACUGACUGG CUCAGCGUGU GCCUACCCUA CGCCGGCAGG CGCGGGUAAC CCGUUGAACC CCAUUCGUG AUGGGGAUCG GGGAUUGCAA UUAUUCCCCA UGAACGAGGA AUUCCCAGUA AGUGCGGGUC AUAAGCUUGC GUUGAUUAAG UCCCUGCCC UUUGUACACA CCGCCCGUCG CUACUACCGA UUGGAUGGUU UAGUGAGGCC CUCGGAUCGG CCCCGCCGGG G UCGGCCCA CGGCCCUGGC GGAGCGCUGA GAAGACGGUC GAACUUGACU AUCUAGAGGA AGUAAAAGUC GUAACAAGGU UU CCGUAGG UGAACCUGCG GAAGGAUCAU UAACG GENBANK: GENBANK: AL353644.34 |
+Macromolecule #3: U3 snoRNA
| Macromolecule | Name: U3 snoRNA / type: rna / ID: 3 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 70.017203 KDa |
| Sequence | String: AAGACUAUAC UUUCAGGGAU CAUUUCUAUA GUGUGUUACU AGAGAAGUUU CUCUGAACGU GUAGAGCACC GAAAACCACG AGGAAGAGA GGUAGCGUUU UCUCCUGAGC GUGAAGCCGG CUUUCUGGCG UUGCUUGGCU GCAACUGCCG UCAGCCAUUG A UGAUCGUU ...String: AAGACUAUAC UUUCAGGGAU CAUUUCUAUA GUGUGUUACU AGAGAAGUUU CUCUGAACGU GUAGAGCACC GAAAACCACG AGGAAGAGA GGUAGCGUUU UCUCCUGAGC GUGAAGCCGG CUUUCUGGCG UUGCUUGGCU GCAACUGCCG UCAGCCAUUG A UGAUCGUU CUUCUCUCCG UAUUGGGGAG UGAGAGGGAG AGAACGCGGU CUGAGUGGU GENBANK: GENBANK: X14945.1 |
+Macromolecule #62: 5' ETS rRNA
| Macromolecule | Name: 5' ETS rRNA / type: rna / ID: 62 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.197572 KDa |
| Sequence | String: AAAAAAAAAA AAAAAAAAAA AA |
+Macromolecule #4: 40S ribosomal protein S18
| Macromolecule | Name: 40S ribosomal protein S18 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 12.671983 KDa |
| Sequence | String: MSLVIPEKFQ HILRVLNTNI DGRRKIAFAI TAIKGVGRRY AHVVLRKADI DLTKRAGELT EDEVERVITI MQNPRQYKIP DWFLNRQ(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK) ...String: MSLVIPEKFQ HILRVLNTNI DGRRKIAFAI TAIKGVGRRY AHVVLRKADI DLTKRAGELT EDEVERVITI MQNPRQYKIP DWFLNRQ(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) |
+Macromolecule #5: 40S ribosomal protein S4 X isoform
| Macromolecule | Name: 40S ribosomal protein S4 X isoform / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 29.654869 KDa |
| Sequence | String: MARGPKKHLK RVAAPKHWML DKLTGVFAPR PSTGPHKLRE CLPLIIFLRN RLKYALTGDE VKKICMQRFI KIDGKVRTDI TYPAGFMDV ISIDKTGENF RLIYDTKGRF AVHRITPEEA KYKLCKVRKI FVGTKGIPHL VTHDARTIRY PDPLIKVNDT I QIDLETGK ...String: MARGPKKHLK RVAAPKHWML DKLTGVFAPR PSTGPHKLRE CLPLIIFLRN RLKYALTGDE VKKICMQRFI KIDGKVRTDI TYPAGFMDV ISIDKTGENF RLIYDTKGRF AVHRITPEEA KYKLCKVRKI FVGTKGIPHL VTHDARTIRY PDPLIKVNDT I QIDLETGK ITDFIKFDTG NLCMVTGGAN LGRIGVITNR ERHPGSFDVV HVKDANGNSF ATRLSNIFVI GKGNKPWISL PR GKGIRLT IAEERDKRLA AKQSSG |
+Macromolecule #6: 40S ribosomal protein S5
| Macromolecule | Name: 40S ribosomal protein S5 / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 22.913453 KDa |
| Sequence | String: MTEWETAAPA VAETPDIKLF GKWSTDDVQI NDISLQDYIA VKEKYAKYLP HSAGRYAAKR FRKAQCPIVE RLTNSMMMHG RNNGKKLMT VRIVKHAFEI IHLLTGENPL QVLVNAIINS GPREDSTRIG RAGTVRRQAV DVSPLRRVNQ AIWLLCTGAR E AAFRNIKT ...String: MTEWETAAPA VAETPDIKLF GKWSTDDVQI NDISLQDYIA VKEKYAKYLP HSAGRYAAKR FRKAQCPIVE RLTNSMMMHG RNNGKKLMT VRIVKHAFEI IHLLTGENPL QVLVNAIINS GPREDSTRIG RAGTVRRQAV DVSPLRRVNQ AIWLLCTGAR E AAFRNIKT IAECLADELI NAAKGSSNSY AIKKKDELER VAKSNR UniProtKB: Small ribosomal subunit protein uS7 |
+Macromolecule #7: 40S ribosomal protein S6
| Macromolecule | Name: 40S ribosomal protein S6 / type: protein_or_peptide / ID: 7 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 28.751906 KDa |
| Sequence | String: MKLNISFPAT GCQKLIEVDD ERKLRTFYEK RMATEVAADA LGEEWKGYVV RISGGNDKQG FPMKQGVLTH GRVRLLLSKG HSCYRPRRT GERKRKSVRG CIVDANLSVL NLVIVKKGEK DIPGLTDTTV PRRLGPKRAS RIRKLFNLSK EDDVRQYVVR K PLNKEGKK ...String: MKLNISFPAT GCQKLIEVDD ERKLRTFYEK RMATEVAADA LGEEWKGYVV RISGGNDKQG FPMKQGVLTH GRVRLLLSKG HSCYRPRRT GERKRKSVRG CIVDANLSVL NLVIVKKGEK DIPGLTDTTV PRRLGPKRAS RIRKLFNLSK EDDVRQYVVR K PLNKEGKK PRTKAPKIQR LVTPRVLQHK RRRIALKKQR TKKNKEEAAE YAKLLAKRMK EAKEKRQEQI AKRRRLSSLR AS TSKSESS QK UniProtKB: Small ribosomal subunit protein eS6 |
+Macromolecule #8: 40S ribosomal protein S7
| Macromolecule | Name: 40S ribosomal protein S7 / type: protein_or_peptide / ID: 8 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 22.168914 KDa |
| Sequence | String: MFSSSAKIVK PNGEKPDEFE SGISQALLEL EMNSDLKAQL RELNITAAKE IEVGGGRKAI IIFVPVPQLK SFQKIQVRLV RELEKKFSG KHVVFIAQRR ILPKPTRKSR TKNKQKRPRS RTLTAVHDAI LEDLVFPSEI VGKRIRVKLD GSRLIKVHLD K AQQNNVEH ...String: MFSSSAKIVK PNGEKPDEFE SGISQALLEL EMNSDLKAQL RELNITAAKE IEVGGGRKAI IIFVPVPQLK SFQKIQVRLV RELEKKFSG KHVVFIAQRR ILPKPTRKSR TKNKQKRPRS RTLTAVHDAI LEDLVFPSEI VGKRIRVKLD GSRLIKVHLD K AQQNNVEH KVETFSGVYK KLTGKDVNFE FPEFQL UniProtKB: Small ribosomal subunit protein eS7 |
+Macromolecule #9: 40S ribosomal protein S8
| Macromolecule | Name: 40S ribosomal protein S8 / type: protein_or_peptide / ID: 9 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 24.263387 KDa |
| Sequence | String: MGISRDNWHK RRKTGGKRKP YHKKRKYELG RPAANTKIGP RRIHTVRVRG GNKKYRALRL DVGNFSWGSE CCTRKTRIID VVYNASNNE LVRTKTLVKN CIVLIDSTPY RQWYESHYAL PLGRKKGAKL TPEEEEILNK KRSKKIQKKY DERKKNAKIS S LLEEQFQQ ...String: MGISRDNWHK RRKTGGKRKP YHKKRKYELG RPAANTKIGP RRIHTVRVRG GNKKYRALRL DVGNFSWGSE CCTRKTRIID VVYNASNNE LVRTKTLVKN CIVLIDSTPY RQWYESHYAL PLGRKKGAKL TPEEEEILNK KRSKKIQKKY DERKKNAKIS S LLEEQFQQ GKLLACIASR PGQCGRADGY VLEGKELEFY LRKIKARKGK UniProtKB: Small ribosomal subunit protein eS8 |
+Macromolecule #10: 40S ribosomal protein S9
| Macromolecule | Name: 40S ribosomal protein S9 / type: protein_or_peptide / ID: 10 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 22.641564 KDa |
| Sequence | String: MPVARSWVCR KTYVTPRRPF EKSRLDQELK LIGEYGLRNK REVWRVKFTL AKIRKAAREL LTLDEKDPRR LFEGNALLRR LVRIGVLDE GKMKLDYILG LKIEDFLERR LQTQVFKLGL AKSIHHARVL IRQRHIRVRK QVVNIPSFIV RLDSQKHIDF S LRSPYGGG ...String: MPVARSWVCR KTYVTPRRPF EKSRLDQELK LIGEYGLRNK REVWRVKFTL AKIRKAAREL LTLDEKDPRR LFEGNALLRR LVRIGVLDE GKMKLDYILG LKIEDFLERR LQTQVFKLGL AKSIHHARVL IRQRHIRVRK QVVNIPSFIV RLDSQKHIDF S LRSPYGGG RPGRVKRKNA KKGQGGAGAG DDEEED UniProtKB: Small ribosomal subunit protein uS4 |
+Macromolecule #11: 40S ribosomal protein S12
| Macromolecule | Name: 40S ribosomal protein S12 / type: protein_or_peptide / ID: 11 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 14.538987 KDa |
| Sequence | String: MAEEGIAAGG VMDVNTALQE VLKTALIHDG LARGIREAAK ALDKRQAHLC VLASNCDEPM YVKLVEALCA EHQINLIKVD DNKKLGEWV GLCKIDREGK PRKVVGCSCV VVKDYGKESQ AKDVIEEYFK CKK UniProtKB: Small ribosomal subunit protein eS12 |
+Macromolecule #12: 40S ribosomal protein S16
| Macromolecule | Name: 40S ribosomal protein S16 / type: protein_or_peptide / ID: 12 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 16.477377 KDa |
| Sequence | String: MPSKGPLQSV QVFGRKKTAT AVAHCKRGNG LIKVNGRPLE MIEPRTLQYK LLEPVLLLGK ERFAGVDIRV RVKGGGHVAQ IYAIRQSIS KALVAYYQKY VDEASKKEIK DILIQYDRTL LVADPRRCES KKFGGPGARA RYQKSYR UniProtKB: Small ribosomal subunit protein uS9 |
+Macromolecule #13: 40S ribosomal protein S11
| Macromolecule | Name: 40S ribosomal protein S11 / type: protein_or_peptide / ID: 13 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 18.468826 KDa |
| Sequence | String: MADIQTERAY QKQPTIFQNK KRVLLGETGK EKLPRYYKNI GLGFKTPKEA IEGTYIDKKC PFTGNVSIRG RILSGVVTKM KMQRTIVIR RDYLHYIRKY NRFEKRHKNM SVHLSPCFRD VQIGDIVTVG ECRPLSKTVR FNVLKVTKAA GTKKQFQKF UniProtKB: Small ribosomal subunit protein uS17 |
+Macromolecule #14: 40S ribosomal protein S24
| Macromolecule | Name: 40S ribosomal protein S24 / type: protein_or_peptide / ID: 14 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 15.463333 KDa |
| Sequence | String: MNDTVTIRTR KFMTNRLLQR KQMVIDVLHP GKATVPKTEI REKLAKMYKT TPDVIFVFGF RTHFGGGKTT GFGMIYDSLD YAKKNEPKH RLARHGLYEK KKTSRKQRKE RKNRMKKVRG TAKANVGAGK KPKE UniProtKB: Small ribosomal subunit protein eS24 |
+Macromolecule #15: 40S ribosomal protein S28
| Macromolecule | Name: 40S ribosomal protein S28 / type: protein_or_peptide / ID: 15 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.855052 KDa |
| Sequence | String: MDTSRVQPIK LARVTKVLGR TGSQGQCTQV RVEFMDDTSR SIIRNVKGPV REGDVLTLLE SEREARRLR UniProtKB: Small ribosomal subunit protein eS28 |
+Macromolecule #16: WD repeat-containing protein 75
| Macromolecule | Name: WD repeat-containing protein 75 / type: protein_or_peptide / ID: 16 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 94.609609 KDa |
| Sequence | String: MVEEENIRVV RCGGSELNFR RAVFSADSKY IFCVSGDFVK VYSTVTEECV HILHGHRNLV TGIQLNPNNH LQLYSCSLDG TIKLWDYID GILIKTFIVG CKLHALFTLA QAEDSVFVIV NKEKPDIFQL VSVKLPKSSS QEVEAKELSF VLDYINQSPK C IAFGNEGV ...String: MVEEENIRVV RCGGSELNFR RAVFSADSKY IFCVSGDFVK VYSTVTEECV HILHGHRNLV TGIQLNPNNH LQLYSCSLDG TIKLWDYID GILIKTFIVG CKLHALFTLA QAEDSVFVIV NKEKPDIFQL VSVKLPKSSS QEVEAKELSF VLDYINQSPK C IAFGNEGV YVAAVREFYL SVYFFKKKTT SRFTLSSSRN KKHAKNNFTC VACHPTEDCI ASGHMDGKIR LWRNFYDDKK YT YTCLHWH HDMVMDLAFS VTGTSLLSGG RESVLVEWRD ATEKNKEFLP RLGATIEHIS VSPAGDLFCT SHSDNKIIII HRN LEASAV IQGLVKDRSI FTGLMIDPRT KALVLNGKPG HLQFYSLQSD KQLYNLDIIQ QEYINDYGLI QIELTKAAFG CFGN WLATV EQRQEKETEL ELQMKLWMYN KKTQGFILNT KINMPHEDCI TALCFCNAEK SEQPTLVTAS KDGYFKVWIL TDDSD IYKK AVGWTCDFVG SYHKYQATNC CFSEDGSLLA VSFEEIVTIW DSVTWELKCT FCQRAGKIRH LCFGRLTCSK YLLGAT ENG ILCCWNLLSC ALEWNAKLNV RVMEPDPNSE NIAAISQSSV GSDLFVFKPS EPRPLYIQKG ISREKVQWGV FVPRDVP ES FTSEAYQWLN RSQFYFLTKS QSLLTFSTKS PEEKLTPTSK QLLAEESLPT TPFYFILGKH RQQQDEKLNE TLENELVQ L PLTENIPAIS ELLHTPAHVL PSAAFLCSMF VNSLLLSKET KSAKEIPEDV DMEEEKESED SDEENDFTEK VQDTSNTGL GEDIIHQLSK SEEKELRKFR KIDYSWIAAL UniProtKB: WD repeat-containing protein 75 |
+Macromolecule #17: Nucleolar protein 11
| Macromolecule | Name: Nucleolar protein 11 / type: protein_or_peptide / ID: 17 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 75.231891 KDa |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) ...String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)VI KWTKFFVVFR HPVLIFIT E KHGNYFAYVQ MFNSRILTKY TLLLGQDENS VIKSFTASVD RKFISLMSLS SDGCIYETLI PIRPADPEKN QSLVKSLLL KAVVSGNARN GVALTALDQD HVAVLGSPLA ASKECLSVWN IKFQTLQTSK ELPQGTSGQL WYYGEHLFML H(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)HVVSH FVNWETPQGC GLGFQNSEQS RRILRRRKIE VSLQPEVPPS KQLLSTIMKD SEKHIEVEV RKFLALKQTP DFHTVIGDTV TGLLERCKAE PSFYPRNCLM QLIQTHVLSY SLCPDLMEIA LKKKDVQLLQ L CLQQFPDI PESVTCACLK IFLSIGDDSL QETDVNMESV FDYSINSVHD EKMEEQTEIL QNGFNPEEDK CNNCDQELNK KP QDETKES TSCPVVQKRA ALLNAILHSA YSETFLLPHL KDIPAQHITL FLKYLYFLYL KCSENATMTL PGIHPPTLNQ IMD WICLLL DANFTVVVMM PEAKRLLINL YKLVKSQISV YSELNKIEVS FRELQKLNQE KNNRGLYSIE VLELF |
+Macromolecule #18: U3 small nucleolar RNA-associated protein 15 homolog
| Macromolecule | Name: U3 small nucleolar RNA-associated protein 15 homolog / type: protein_or_peptide / ID: 18 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 58.503156 KDa |
| Sequence | String: MAGYKPVAIQ TYPILGEKIT QDTLYWNNYK TPVQIKEFGA VSKVDFSPQP PYNYAVTASS RIHIYGRYSQ EPIKTFSRFK DTAYCATFR QDGRLLVAGS EDGGVQLFDI SGRAPLRQFE GHTKAVHTVD FTADKYHVVS GADDYTVKLW DIPNSKEILT F KEHSDYVR ...String: MAGYKPVAIQ TYPILGEKIT QDTLYWNNYK TPVQIKEFGA VSKVDFSPQP PYNYAVTASS RIHIYGRYSQ EPIKTFSRFK DTAYCATFR QDGRLLVAGS EDGGVQLFDI SGRAPLRQFE GHTKAVHTVD FTADKYHVVS GADDYTVKLW DIPNSKEILT F KEHSDYVR CGCASKLNPD LFITGSYDHT VKMFDARTSE SVLSVEHGQP VESVLLFPSG GLLVSAGGRY VKVWDMLKGG QL LVSLKNH HKTVTCLCLS SSGQRLLSGS LDRKVKVYST TSYKVVHSFD YAASILSLAL AHEDETIVVG MTNGILSVKH RKS EAKKES LPRRRRPAYR TFIKGKNYMK QRDDILINRP AKKHLELYDR DLKHFRISKA LDRVLDPTCT IKTPEITVSI IKEL NRRGV LANALAGRDE KEISHVLNFL IRNLSQPRFA PVLINAAEII IDIYLPVIGQ SPVVDKKFLL LQGLVEKEID YQREL LETL GMMDMLFATM RRKEGTSVLE HTSDGFPENK KIES UniProtKB: U3 small nucleolar RNA-associated protein 15 homolog |
+Macromolecule #19: WD repeat-containing protein 43
| Macromolecule | Name: WD repeat-containing protein 43 / type: protein_or_peptide / ID: 19 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 74.985578 KDa |
| Sequence | String: MAAGGGGSCD PLAPAGVPCA FSPHSQAYFA LASTDGHLRV WETANNRLHQ EYVPSAHLSG TCTCLAWAPA RLQAKESPQR KKRKSEAVG MSNQTDLLAL GTAVGSILLY STVKGELHSK LISGGHDNRV NCIQWHQDSG CLYSCSDDKH IVEWNVQTCK V KCKWKGDN ...String: MAAGGGGSCD PLAPAGVPCA FSPHSQAYFA LASTDGHLRV WETANNRLHQ EYVPSAHLSG TCTCLAWAPA RLQAKESPQR KKRKSEAVG MSNQTDLLAL GTAVGSILLY STVKGELHSK LISGGHDNRV NCIQWHQDSG CLYSCSDDKH IVEWNVQTCK V KCKWKGDN SSVSSLCISP DGKMLLSAGR TIKLWVLETK EVYRHFTGHA TPVSSLMFTT IRPPNESQPF DGITGLYFLS GA VHDRLLN VWQVRSENKE KSAVMSFTVT DEPVYIDLTL SENKEEPVKL AVVCRDGQVH LFEHILNGYC KKPLTSNCTI QIA TPGKGK KSTPKPIPIL AAGFCSDKMS LLLVYGSWFQ PTIERVALNS REPHMCLVRD ISNCWAPKVE TAITKVRTPV MNSE AKVLV PGIPGHHAAI KPAPPQTEQV ESKRKSGGNE VSIEERLGAM DIDTHKKGKE DLQTNSFPVL LTQGLESNDF EMLNK VLQT RNVNLIKKTV LRMPLHTIIP LLQELTKRLQ GHPNSAVLMV QWLKCVLTVH ASYLSTLPDL VPQLGTLYQL MESRVK TFQ KLSHLHGKLI LLITQVTASE KTKGATSPGQ KAKLVYEEES SEEESDDEIA DKDSEDNWDE DEEESESEKD EDVEEED ED AEGKDEENGE DRDTASEKEL NGDSDLDPEN ESEEE UniProtKB: WD repeat-containing protein 43 |
+Macromolecule #20: U3 small nucleolar RNA-associated protein 4 homolog
| Macromolecule | Name: U3 small nucleolar RNA-associated protein 4 homolog / type: protein_or_peptide / ID: 20 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 76.993055 KDa |
| Sequence | String: MGEFKVHRVR FFNYVPSGIR CVAYNNQSNR LAVSRTDGTV EIYNLSANYF QEKFFPGHES RATEALCWAE GQRLFSAGLN GEIMEYDLQ ALNIKYAMDA FGGPIWSMAA SPSGSQLLVG CEDGSVKLFQ ITPDKIQFER NFDRQKSRIL SLSWHPSGTH I AAGSIDYI ...String: MGEFKVHRVR FFNYVPSGIR CVAYNNQSNR LAVSRTDGTV EIYNLSANYF QEKFFPGHES RATEALCWAE GQRLFSAGLN GEIMEYDLQ ALNIKYAMDA FGGPIWSMAA SPSGSQLLVG CEDGSVKLFQ ITPDKIQFER NFDRQKSRIL SLSWHPSGTH I AAGSIDYI SVFDVKSGSA VHKMIVDRQY MGVSKRKCIV WGVAFLSDGT IISVDSAGKV QFWDSATGTL VKSHLIANAD VQ SIAVADQ EDSFVVGTAE GTVFHFQLVP VTSNSSEKQW VRTKPFQHHT HDVRTVAHSP TALISGGTDT HLVFRPLMEK VEV KNYDAA LRKITFPHRC LISCSKKRQL LLFQFAHHLE LWRLGSTVAT GKNGDTLPLS KNADHLLHLK TKGPENIICS CISP CGSWI AYSTVSRFFL YRLNYEHDNI SLKRVSKMPA FLRSALQILF SEDSTKLFVA SNQGALHIVQ LSGGSFKHLH AFQPQ SGTV EAMCLLAVSP DGNWLAASGT SAGVHVYNVK QLKLHCTVPA YNFPVTAMAI APNTNNLVIA HSDQQVFEYS IPDKQY TDW SRTVQKQGFH HLWLQRDTPI THISFHPKRP MHILLHDAYM FCIIDKSLPL PNDKTLLYNP FPPTNESDVI RRRTAHA FK ISKIYKPLLF MDLLDERTLV AVERPLDDII AQLPPPIKKK KFGT UniProtKB: U3 small nucleolar RNA-associated protein 4 homolog |
+Macromolecule #21: Periodic tryptophan protein 2 homolog
| Macromolecule | Name: Periodic tryptophan protein 2 homolog / type: protein_or_peptide / ID: 21 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 102.574977 KDa |
| Sequence | String: MKFAYRFSNL LGTVYRRGNL NFTCDGNSVI SPVGNRVTVF DLKNNKSDTL PLATRYNVKC VGLSPDGRLA IIVDEGGDAL LVSLVCRSV LHHFHFKGSV HSVSFSPDGR KFVVTKGNIA QMYHAPGKKR EFNAFVLDKT YFGPYDETTC IDWTDDSRCF V VGSKDMST ...String: MKFAYRFSNL LGTVYRRGNL NFTCDGNSVI SPVGNRVTVF DLKNNKSDTL PLATRYNVKC VGLSPDGRLA IIVDEGGDAL LVSLVCRSV LHHFHFKGSV HSVSFSPDGR KFVVTKGNIA QMYHAPGKKR EFNAFVLDKT YFGPYDETTC IDWTDDSRCF V VGSKDMST WVFGAERWDN LIYYALGGHK DAIVACFFES NSLDLYSLSQ DGVLCMWQCD TPPEGLRLKP PAGWKADLLQ RE EEEEEEE DQEGDRETTI RGKATPAEEE KTGKVKYSRL AKYFFNKEGD FNNLTAAAFH KKSHLLVTGF ASGIFHLHEL PEF NLIHSL SISDQSIASV AINSSGDWIA FGCSGLGQLL VWEWQSESYV LKQQGHFNSM VALAYSPDGQ YIVTGGDDGK VKVW NTLSG FCFVTFTEHS SGVTGVTFTA TGYVVVTSSM DGTVRAFDLH RYRNFRTFTS PRPTQFSCVA VDASGEIVSA GAQDS FEIF VWSMQTGRLL DVLSGHEGPI SGLCFNPMKS VLASASWDKT VRLWDMFDSW RTKETLALTS DALAVTFRPD GAELAV ATL NSQITFWDPE NAVQTGSIEG RHDLKTGRKE LDKITAKHAA KGKAFTALCY SADGHSILAG GMSKFVCIYH VREQILM KR FEISCNLSLD AMEEFLNRRK MTEFGNLALI DQDAGQEDGV AIPLPGVRKG DMSSRHFKPE IRVTSLRFSP TGRCWAAT T TEGLLIYSLD TRVLFDPFEL DTSVTPGRVR EALRQQDFTR AILMALRLNE SKLVQEALEA VPRGEIEVVT SSLPELYVE KVLEFLASSF EVSRHLEFYL LWTHKLLMLH GQKLKSRAGT LLPVIQFLQK SIQRHLDDLS KLCSWNHYNM QYALAVSKQR GTKRSLDPL GSEEEAEASE DDSLHLLGGG GRDSEEEMLA UniProtKB: Periodic tryptophan protein 2 homolog |
+Macromolecule #22: U3 small nucleolar RNA-associated protein 6 homolog
| Macromolecule | Name: U3 small nucleolar RNA-associated protein 6 homolog / type: protein_or_peptide / ID: 22 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 70.297891 KDa |
| Sequence | String: MAEIIQERIE DRLPELEQLE RIGLFSHAEI KAIIKKASDL EYKIQRRTLF KEDFINYVQY EINLLELIQR RRTRIGYSFK KDEIENSIV HRVQGVFQRA SAKWKDDVQL WLSYVAFCKK WATKTRLSKV FSAMLAIHSN KPALWIMAAK WEMEDRLSSE S ARQLFLRA ...String: MAEIIQERIE DRLPELEQLE RIGLFSHAEI KAIIKKASDL EYKIQRRTLF KEDFINYVQY EINLLELIQR RRTRIGYSFK KDEIENSIV HRVQGVFQRA SAKWKDDVQL WLSYVAFCKK WATKTRLSKV FSAMLAIHSN KPALWIMAAK WEMEDRLSSE S ARQLFLRA LRFHPECPKL YKEYFRMELM HAEKLRKEKE EFEKASMDVE NPDYSEEILK GELAWIIYKN SVSIIKGAEF HV SLLSIAQ LFDFAKDLQK EIYDDLQALH TDDPLTWDYV ARRELEIESQ TEEQPTTKQA KAVEVGRKEE RCCAVYEEAV KTL PTEAMW KCYITFCLER FTKKSNSGFL RGKRLERTMT VFRKAHELKL LSECQYKQLS VSLLCYNFLR EALEVAVAGT ELFR DSGTM WQLKLQVLIE SKSPDIAMLF EEAFVHLKPQ VCLPLWISWA EWSEGAKSQE DTEAVFKKAL LAVIGADSVT LKNKY LDWA YRSGGYKKAR AVFKSLQESR PFSVDFFRKM IQFEKEQESC NMANIREYYE RALREFGSAD SDLWMDYMKE ELNHPL GRP ENCGQIYWRA MKMLQGESAE AFVAKHAMHQ TGHL UniProtKB: U3 small nucleolar RNA-associated protein 6 homolog |
+Macromolecule #23: WD repeat-containing protein 3
| Macromolecule | Name: WD repeat-containing protein 3 / type: protein_or_peptide / ID: 23 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 106.248 KDa |
| Sequence | String: MGLTKQYLRY VASAVFGVIG SQKGNIVFVT LRGEKGRYVA VPACEHVFIW DLRKGEKILI LQGLKQEVTC LCPSPDGLHL AVGYEDGSI RIFSLLSGEG NVTFNGHKAA ITTLKYDQLG GRLASGSKDT DIIVWDVINE SGLYRLKGHK DAITQALFLR E KNLLVTSG ...String: MGLTKQYLRY VASAVFGVIG SQKGNIVFVT LRGEKGRYVA VPACEHVFIW DLRKGEKILI LQGLKQEVTC LCPSPDGLHL AVGYEDGSI RIFSLLSGEG NVTFNGHKAA ITTLKYDQLG GRLASGSKDT DIIVWDVINE SGLYRLKGHK DAITQALFLR E KNLLVTSG KDTMVKWWDL DTQHCFKTMV GHRTEVWGLV LLSEEKRLIT GASDSELRVW DIAYLQEIED PEEPDPKKIK GS SPGIQDT LEAEDGAFET DEAPEDRILS CRKAGSIMRE GRDRVVNLAV DKTGRILACH GTDSVLELFC ILSKKEIQKK MDK KMKKAR KKAKLHSSKG EEEDPEVNVE MSLQDEIQRV TNIKTSAKIK SFDLIHSPHG ELKAVFLLQN NLVELYSLNP SLPT PQPVR TSRITIGGHR SDVRTLSFSS DNIAVLSAAA DSIKIWNRST LQCIRTMTCE YALCSFFVPG DRQVVIGTKT GKLQL YDLA SGNLLETIDA HDGALWSMSL SPDQRGFVTG GADKSVKFWD FELVKDENST QKRLSVKQTR TLQLDEDVLC VSYSPN QKL LAVSLLDCTV KIFYVDTLKF FLSLYGHKLP VICMDISHDG ALIATGSADR NVKIWGLDFG DCHKSLFAHD DSVMYLQ FV PKSHLFFTAG KDHKIKQWDA DKFEHIQTLE GHHQEIWCLA VSPSGDYVVS SSHDKSLRLW ERTREPLILE EEREMERE A EYEESVAKED QPAVPGETQG DSYFTGKKTI ETVKAAERIM EAIELYREET AKMKEHKAIC KAAGKEVPLP SNPILMAYG SISPSAYVLE IFKGIKSSEL EESLLVLPFS YVPDILKLFN EFIQLGSDVE LICRCLFFLL RIHFGQITSN QMLVPVIEKL RETTISKVS QVRDVIGFNM AGLDYLKREC EAKSEVMFFA DATSHLEEKK RKRKKREKLI LTLT UniProtKB: WD repeat-containing protein 3 |
+Macromolecule #24: U3 small nucleolar RNA-associated protein 18 homolog
| Macromolecule | Name: U3 small nucleolar RNA-associated protein 18 homolog / type: protein_or_peptide / ID: 24 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 62.097547 KDa |
| Sequence | String: MPPERRRRMK LDRRTGAKPK RKPGMRPDWK AGAGPGGPPQ KPAPSSQRKP PARPSAAAAA IAVAAAEEER RLRQRNRLRL EEDKPAVER CLEELVFGDV ENDEDALLRR LRGPRVQEHE DSGDSEVENE AKGNFPPQKK PVWVDEEDED EEMVDMMNNR F RKDMMKNA ...String: MPPERRRRMK LDRRTGAKPK RKPGMRPDWK AGAGPGGPPQ KPAPSSQRKP PARPSAAAAA IAVAAAEEER RLRQRNRLRL EEDKPAVER CLEELVFGDV ENDEDALLRR LRGPRVQEHE DSGDSEVENE AKGNFPPQKK PVWVDEEDED EEMVDMMNNR F RKDMMKNA SESKLSKDNL KKRLKEEFQH AMGGVPAWAE TTKRKTSSDD ESEEDEDDLL QRTGNFISTS TSLPRGILKM KN CQHANAE RPTVARISSV QFHPGAQIVM VAGLDNAVSL FQVDGKTNPK IQSIYLERFP IFKACFSANG EEVLATSTHS KVL YVYDML AGKLIPVHQV RGLKEKIVRS FEVSPDGSFL LINGIAGYLH LLAMKTKELI GSMKINGRVA ASTFSSDSKK VYAS SGDGE VYVWDVNSRK CLNRFVDEGS LYGLSIATSR NGQYVACGSN CGVVNIYNQD SCLQETNPKP IKAIMNLVTG VTSLT FNPT TEILAIASEK MKEAVRLVHL PSCTVFSNFP VIKNKNISHV HTMDFSPRSG YFALGNEKGK ALMYRLHHYS DF UniProtKB: U3 small nucleolar RNA-associated protein 18 homolog |
+Macromolecule #25: WD repeat-containing protein 36
| Macromolecule | Name: WD repeat-containing protein 36 / type: protein_or_peptide / ID: 25 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 105.443 KDa |
| Sequence | String: MCCTEGSLRK RDSQRAPEAV LCLQLWQRTV PLDTLKGLGT CFPSGPELRG AGIAAAMERA SERRTASALF AGFRALGLFS NDIPHVVRF SALKRRFYVT TCVGKSFHTY DVQKLSLVAV SNSVPQDICC MAADGRLVFA AYGNVFSAFA RNKEIVHTFK G HKAEIHFL ...String: MCCTEGSLRK RDSQRAPEAV LCLQLWQRTV PLDTLKGLGT CFPSGPELRG AGIAAAMERA SERRTASALF AGFRALGLFS NDIPHVVRF SALKRRFYVT TCVGKSFHTY DVQKLSLVAV SNSVPQDICC MAADGRLVFA AYGNVFSAFA RNKEIVHTFK G HKAEIHFL QPFGDHIISV DTDGILIIWH IYSEEEYLQL TFDKSVFKIS AILHPSTYLN KILLGSEQGS LQLWNVKSNK LL YTFPGWK VGVTALQQAP AVDVVAIGLM SGQVIIHNIK FNETLMKFRQ DWGPITSISF RTDGHPVMAA GSPCGHIGLW DLE DKKLIN QMRNAHSTAI AGLTFLHREP LLVTNGADNA LRIWIFDGPT GEGRLLRFRM GHSAPLTNIR YYGQNGQQIL SASQ DGTLQ SFSTVHEKFN KSLGHGLINK KRVKRKGLQN TMSVRLPPIT KFAAEEARES DWDGIIACHQ GKLSCSTWNY QKSTI GAYF LKPKELKKDD ITATAVDITS CGNFAVIGLS SGTVDVYNMQ SGIHRGSFGK DQAHKGSVRG VAVDGLNQLT VTTGSE GLL KFWNFKNKIL IHSVSLSSSP NIMLLHRDSG ILGLALDDFS ISVLDIETRK IVREFSGHQG QINDMAFSPD GRWLISA AM DCSIRTWDLP SGCLIDCFLL DSAPLNVSMS PTGDFLATSH VDHLGIYLWS NISLYSVVSL RPLPADYVPS IVMLPGTC Q TQDVEVSEET VEPSDELIEY DSPEQLNEQL VTLSLLPESR WKNLLNLDVI KKKNKPKEPP KVPKSAPFFI PTIPGLVPR YAAPEQNNDP QQSKVVNLGV LAQKSDFCLK LEEGLVNNKY DTALNLLKES GPSGIETELR SLSPDCGGSI EVMQSFLKMI GMMLDRKRD FELAQAYLAL FLKLHLKMLP SEPVLLEEIT NLSSQVEENW THLQSLFNQS MCILNYLKSA LL UniProtKB: WD repeat-containing protein 36 |
+Macromolecule #26: DDB1- and CUL4-associated factor 13
| Macromolecule | Name: DDB1- and CUL4-associated factor 13 / type: protein_or_peptide / ID: 26 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 51.489266 KDa |
| Sequence | String: MKVKMLSRNP DNYVRETKLD LQRVPRNYDP ALHPFEVPRE YIRALNATKL ERVFAKPFLA SLDGHRDGVN CLAKHPEKLA TVLSGACDG EVRIWNLTQR NCIRTIQAHE GFVRGICTRF CGTSFFTVGD DKTVKQWKMD GPGYGDEEEP LHTILGKTVY T GIDHHWKE ...String: MKVKMLSRNP DNYVRETKLD LQRVPRNYDP ALHPFEVPRE YIRALNATKL ERVFAKPFLA SLDGHRDGVN CLAKHPEKLA TVLSGACDG EVRIWNLTQR NCIRTIQAHE GFVRGICTRF CGTSFFTVGD DKTVKQWKMD GPGYGDEEEP LHTILGKTVY T GIDHHWKE AVFATCGQQV DIWDEQRTNP ICSMTWGFDS ISSVKFNPIE TFLLGSCASD RNIVLYDMRQ ATPLKKVILD MR TNTICWN PMEAFIFTAA NEDYNLYTFD MRALDTPVMV HMDHVSAVLD VDYSPTGKEF VSASFDKSIR IFPVDKSRSR EVY HTKRMQ HVICVKWTSD SKYIMCGSDE MNIRLWKANA SEKLGVLTSR EKAAKDYNQK LKEKFQHYPH IKRIARHRHL PKSI YSQIQ EQRIMKEARR RKEVNRIKHS KPGSVPLVSE KKKHVVAVVK UniProtKB: DDB1- and CUL4-associated factor 13 |
+Macromolecule #27: WD repeat-containing protein 46
| Macromolecule | Name: WD repeat-containing protein 46 / type: protein_or_peptide / ID: 27 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 68.189172 KDa |
| Sequence | String: METAPKPGKD VPPKKDKLQT KRKKPRRYWE EETVPTTAGA SPGPPRNKKN RELRPQRPKN AYILKKSRIS KKPQVPKKPR EWKNPESQR GLSGTQDPFP GPAPVPVEVV QKFCRIDKSR KLPHSKAKTR SRLEVAEAEE EETSIKAARS ELLLAEEPGF L EGEDGEDT ...String: METAPKPGKD VPPKKDKLQT KRKKPRRYWE EETVPTTAGA SPGPPRNKKN RELRPQRPKN AYILKKSRIS KKPQVPKKPR EWKNPESQR GLSGTQDPFP GPAPVPVEVV QKFCRIDKSR KLPHSKAKTR SRLEVAEAEE EETSIKAARS ELLLAEEPGF L EGEDGEDT AKICQADIVE AVDIASAAKH FDLNLRQFGP YRLNYSRTGR HLAFGGRRGH VAALDWVTKK LMCEINVMEA VR DIRFLHS EALLAVAQNR WLHIYDNQGI ELHCIRRCDR VTRLEFLPFH FLLATASETG FLTYLDVSVG KIVAALNARA GRL DVMSQN PYNAVIHLGH SNGTVSLWSP AMKEPLAKIL CHRGGVRAVA VDSTGTYMAT SGLDHQLKIF DLRGTYQPLS TRTL PHGAG HLAFSQRGLL VAGMGDVVNI WAGQGKASPP SLEQPYLTHR LSGPVHGLQF CPFEDVLGVG HTGGITSMLV PGAGE PNFD GLESNPYRSR KQRQEWEVKA LLEKVPAELI CLDPRALAEV DVISLEQGKK EQIERLGYDP QAKAPFQPKP KQKGRS STA SLVKRKRKVM DEEHRDKVRQ SLQQQHHKEA KAKPTGARPS ALDRFVR UniProtKB: WD repeat-containing protein 46 |
+Macromolecule #28: U3 small nucleolar ribonucleoprotein protein IMP3
| Macromolecule | Name: U3 small nucleolar ribonucleoprotein protein IMP3 / type: protein_or_peptide / ID: 28 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 21.889227 KDa |
| Sequence | String: MVRKLKFHEQ KLLKQVDFLN WEVTDHNLHE LRVLRRYRLQ RREDYTRYNQ LSRAVRELAR RLRDLPERDQ FRVRASAALL DKLYALGLV PTRGSLELCD FVTASSFCRR RLPTVLLKLR MAQHLQAAVA FVEQGHVRVG PDVVTDPAFL VTRSMEDFVT W VDSSKIKR HVLEYNEERD DFDLEA UniProtKB: U3 small nucleolar ribonucleoprotein protein IMP3 |
+Macromolecule #29: U3 small nucleolar ribonucleoprotein protein MPP10
| Macromolecule | Name: U3 small nucleolar ribonucleoprotein protein MPP10 / type: protein_or_peptide / ID: 29 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 78.988805 KDa |
| Sequence | String: MAPQVWRRRT LERCLTEVGK ATGRPECFLT IQEGLASKFT SLTKVLYDFN KILENGRIHG SPLQKLVIEN FDDEQIWQQL ELQNEPILQ YFQNAVSETI NDEDISLLPE SEEQEREEDG SEIEADDKED LEDLEEEEVS DMGNDDPEMG ERAENSSKSD L RKSPVFSD ...String: MAPQVWRRRT LERCLTEVGK ATGRPECFLT IQEGLASKFT SLTKVLYDFN KILENGRIHG SPLQKLVIEN FDDEQIWQQL ELQNEPILQ YFQNAVSETI NDEDISLLPE SEEQEREEDG SEIEADDKED LEDLEEEEVS DMGNDDPEMG ERAENSSKSD L RKSPVFSD EDSDLDFDIS KLEQQSKVQN KGQGKPREKS IVDDKFFKLS EMEAYLENIE KEEERKDDND EEEEDIDFFE DI DSDEDEG GLFGSKKLKS GKSSRNLKYK DFFDPVESDE DITNVHDDEL DSNKEDDEIA EEEAEELSIS ETDEDDDLQE NED NKQHKE SLKRVTFALP DDAETEDTGV LNVKKNSDEV KSSFEKRQEK MNEKIASLEK ELLEKKPWQL QGEVTAQKRP ENSL LEETL HFDHAVRMAP VITEETTLQL EDIIKQRIRD QAWDDVVRKE KPKEDAYEYK KRLTLDHEKS KLSLAEIYEQ EYIKL NQQK TAEEENPEHV EIQKMMDSLF LKLDALSNFH FIPKPPVPEI KVVSNLPAIT MEEVAPVSVS DAALLAPEEI KEKNKA GDI KTAAEKTATD KKRERRKKKY QKRMKIKEKE KRRKLLEKSS VDQAGKYSKT VASEKLKQLT KTGKASFIKD EGKDKAL KS SQAFFSKLQD QVKMQINDAK KTEKKKKKRQ DISVHKLKL UniProtKB: U3 small nucleolar ribonucleoprotein protein MPP10 |
+Macromolecule #30: Something about silencing protein 10
| Macromolecule | Name: Something about silencing protein 10 / type: protein_or_peptide / ID: 30 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 54.652023 KDa |
| Sequence | String: MVGRSRRRGA AKWAAVRAKA GPTLTDENGD DLGLPPSPGD TSYYQDQVDD FHEARSRAAL AKGWNEVQSG DEEDGEEEEE EVLALDMDD EDDEDGGNAG EEEEEENADD DGGSSVQSEA EASVDPSLSW GQRKKLYYDT DYGSKSRGRQ SQQEAEEEER E EEEEAQII ...String: MVGRSRRRGA AKWAAVRAKA GPTLTDENGD DLGLPPSPGD TSYYQDQVDD FHEARSRAAL AKGWNEVQSG DEEDGEEEEE EVLALDMDD EDDEDGGNAG EEEEEENADD DGGSSVQSEA EASVDPSLSW GQRKKLYYDT DYGSKSRGRQ SQQEAEEEER E EEEEAQII QRRLAQALQE DDFGVAWVEA FAKPVPQVDE AETRVVKDLA KVSVKEKLKM LRKESPELLE LIEDLKVKLT EV KDELEPL LELVEQGIIP PGKGSQYLRT KYNLYLNYCS NISFYLILKA RRVPAHGHPV IERLVTYRNL INKLSVVDQK LSS EIRHLL TLKDDAVKKE LIPKAKSTKP KPKSVSKTSA AACAVTDLSD DSDFDEKAKL KYYKEIEDRQ KLKRKKEENS TEEQ ALEDQ NAKRAITYQI AKNRGLTPRR KKIDRNPRVK HREKFRRAKI RRRGQVREVR KEEQRYSGEL SGIRAGVKKS IKLK UniProtKB: Something about silencing protein 10 |
+Macromolecule #31: Nucleolar protein 7
| Macromolecule | Name: Nucleolar protein 7 / type: protein_or_peptide / ID: 31 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 29.483512 KDa |
| Sequence | String: MVQLRPRASR APASAEAMVD EGQLASEEEE AEHGLLLGQP SSGAAAEPLE EDEEGDDEFD DEAPEELTFA SAQAEAREEE RRVRETVRR DKTLLKEKRK RREELFIEQK KRKLLPDTIL EKLTTASQTN IKKSPGKVKE VNLQKKNEDC EKGNDSKKVK V QKVQSVSQ ...String: MVQLRPRASR APASAEAMVD EGQLASEEEE AEHGLLLGQP SSGAAAEPLE EDEEGDDEFD DEAPEELTFA SAQAEAREEE RRVRETVRR DKTLLKEKRK RREELFIEQK KRKLLPDTIL EKLTTASQTN IKKSPGKVKE VNLQKKNEDC EKGNDSKKVK V QKVQSVSQ NKSYLAVRLK DQDLRDSRQQ AAQAFIHNSL YGPGTNRTTV NKFLSLANKR LPVKRAAVQF LNNAWGIQKK QN AKRFKRR WMVRKMKTKK UniProtKB: U3 small nucleolar RNA-associated protein NOL7 |
+Macromolecule #32: Uncharacterized protein C1orf131
| Macromolecule | Name: Uncharacterized protein C1orf131 / type: protein_or_peptide / ID: 32 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 32.673367 KDa |
| Sequence | String: MRVDSSADPT MSQEQGPGSS TPPSSPTLLD ALLQNLYDFG GTEGETEQKK IIKKRENKKR DVMASAALAA EPSPLPGSLI RGQRKSASS FFKELREERH CAPSGTPTGP EILAAAVPPS SLKNNREQVE VVEFHSNKKR KLTPDHNKNT KQANPSVLER D VDTQEFNL ...String: MRVDSSADPT MSQEQGPGSS TPPSSPTLLD ALLQNLYDFG GTEGETEQKK IIKKRENKKR DVMASAALAA EPSPLPGSLI RGQRKSASS FFKELREERH CAPSGTPTGP EILAAAVPPS SLKNNREQVE VVEFHSNKKR KLTPDHNKNT KQANPSVLER D VDTQEFNL EKARLEVHRF GITGYGKGKE RILEQERAIM LGAKPPKKSY VNYKVLQEQI KEKKAAKEEE KRLAQETDIF KK KKRKGQE DRKSKKKSAP SILSNGRIGQ VGKFKNGTLI LSPVDIKKIN SSRVAK UniProtKB: 40S small subunit processome assembly factor 1 |
+Macromolecule #33: 40S ribosomal protein S13
| Macromolecule | Name: 40S ribosomal protein S13 / type: protein_or_peptide / ID: 33 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 17.259389 KDa |
| Sequence | String: MGRMHAPGKG LSQSALPYRR SVPTWLKLTS DDVKEQIYKL AKKGLTPSQI GVILRDSHGV AQVRFVTGNK ILRILKSKGL APDLPEDLY HLIKKAVAVR KHLERNRKDK DAKFRLILIE SRIHRLARYY KTKRVLPPNW KYESSTASAL VA UniProtKB: Small ribosomal subunit protein uS15 |
+Macromolecule #34: 40S ribosomal protein S14
| Macromolecule | Name: 40S ribosomal protein S14 / type: protein_or_peptide / ID: 34 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 16.302772 KDa |
| Sequence | String: MAPRKGKEKK EEQVISLGPQ VAEGENVFGV CHIFASFNDT FVHVTDLSGK ETICRVTGGM KVKADRDESS PYAAMLAAQD VAQRCKELG ITALHIKLRA TGGNRTKTPG PGAQSALRAL ARSGMKIGRI EDVTPIPSDS TRRKGGRRGR RL UniProtKB: Small ribosomal subunit protein uS11 |
+Macromolecule #35: RNA cytidine acetyltransferase
| Macromolecule | Name: RNA cytidine acetyltransferase / type: protein_or_peptide / ID: 35 / Number of copies: 2 / Enantiomer: LEVO EC number: Transferases; Acyltransferases; Transferring groups other than aminoacyl groups |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 115.882781 KDa |
| Sequence | String: MHRKKVDNRI RILIENGVAE RQRSLFVVVG DRGKDQVVIL HHMLSKATVK ARPSVLWCYK KELGFSSHRK KRMRQLQKKI KNGTLNIKQ DDPFELFIAA TNIRYCYYNE THKILGNTFG MCVLQDFEAL TPNLLARTVE TVEGGGLVVI LLRTMNSLKQ L YTVTMDVH ...String: MHRKKVDNRI RILIENGVAE RQRSLFVVVG DRGKDQVVIL HHMLSKATVK ARPSVLWCYK KELGFSSHRK KRMRQLQKKI KNGTLNIKQ DDPFELFIAA TNIRYCYYNE THKILGNTFG MCVLQDFEAL TPNLLARTVE TVEGGGLVVI LLRTMNSLKQ L YTVTMDVH SRYRTEAHQD VVGRFNERFI LSLASCKKCL VIDDQLNILP ISSHVATMEA LPPQTPDESL GPSDLELREL KE SLQDTQP VGVLVDCCKT LDQAKAVLKF IEGISEKTLR STVALTAARG RGKSAALGLA IAGAVAFGYS NIFVTSPSPD NLH TLFEFV FKGFDALQYQ EHLDYEIIQS LNPEFNKAVI RVNVFREHRQ TIQYIHPADA VKLGQAELVV IDEAAAIPLP LVKS LLGPY LVFMASTING YEGTGRSLSL KLIQQLRQQS AQSQVSTTAE NKTTTTARLA SARTLYEVSL QESIRYAPGD AVEKW LNDL LCLDCLNITR IVSGCPLPEA CELYYVNRDT LFCYHKASEV FLQRLMALYV ASHYKNSPND LQMLSDAPAH HLFCLL PPV PPTQNALPEV LAVIQVCLEG EISRQSILNS LSRGKKASGD LIPWTVSEQF QDPDFGGLSG GRVVRIAVHP DYQGMGY GS RALQLLQMYY EGRFPCLEEK VLETPQEIHT VSSEAVSLLE EVITPRKDLP PLLLKLNERP AERLDYLGVS YGLTPRLL K FWKRAGFVPV YLRQTPNDLT GEHSCIMLKT LTDEDEADQG GWLAAFWKDF RRRFLALLSY QFSTFSPSLA LNIIQNRNM GKPAQPALSR EELEALFLPY DLKRLEMYSR NMVDYHLIMD MIPAISRIYF LNQLGDLALS AAQSALLLGI GLQHKSVDQL EKEIELPSG QLMGLFNRII RKVVKLFNEV QEKAIEEQMV AAKDVVMEPT MKTLSDDLDE AAKEFQEKHK KEVGKLKSMD L SEYIIRGD DEEWNEVLNK AGPNASIISL KSDKKRKLEA KQEPKQSKKL KNRETKNKKD MKLKRKK UniProtKB: RNA cytidine acetyltransferase |
+Macromolecule #36: 40S ribosomal protein S3a
| Macromolecule | Name: 40S ribosomal protein S3a / type: protein_or_peptide / ID: 36 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 30.002061 KDa |
| Sequence | String: MAVGKNKRLT KGGKKGAKKK VVDPFSKKDW YDVKAPAMFN IRNIGKTLVT RTQGTKIASD GLKGRVFEVS LADLQNDEVA FRKFKLITE DVQGKNCLTN FHGMDLTRDK MCSMVKKWQT MIEAHVDVKT TDGYLLRLFC VGFTKKRNNQ IRKTSYAQHQ Q VRQIRKKM ...String: MAVGKNKRLT KGGKKGAKKK VVDPFSKKDW YDVKAPAMFN IRNIGKTLVT RTQGTKIASD GLKGRVFEVS LADLQNDEVA FRKFKLITE DVQGKNCLTN FHGMDLTRDK MCSMVKKWQT MIEAHVDVKT TDGYLLRLFC VGFTKKRNNQ IRKTSYAQHQ Q VRQIRKKM MEIMTREVQT NDLKEVVNKL IPDSIGKDIE KACQSIYPLH DVFVRKVKML KKPKFELGKL MELHGEGSSS GK ATGDETG AKVERADGYE PPVQESV UniProtKB: Small ribosomal subunit protein eS1 |
+Macromolecule #37: Protein AATF
| Macromolecule | Name: Protein AATF / type: protein_or_peptide / ID: 37 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 63.224887 KDa |
| Sequence | String: MAGPQPLALQ LEQLLNPRPS EADPEADPEE ATAARVIDRF DEGEDGEGDF LVVGSIRKLA SASLLDTDKR YCGKTTSRKA WNEDHWEQT LPGSSDEEIS DEEGSGDEDS EGLGLEEYDE DDLGAAEEQE CGDHRESKKS RSHSAKTPGF SVQSISDFEK F TKGMDDLG ...String: MAGPQPLALQ LEQLLNPRPS EADPEADPEE ATAARVIDRF DEGEDGEGDF LVVGSIRKLA SASLLDTDKR YCGKTTSRKA WNEDHWEQT LPGSSDEEIS DEEGSGDEDS EGLGLEEYDE DDLGAAEEQE CGDHRESKKS RSHSAKTPGF SVQSISDFEK F TKGMDDLG SSEEEEDEES GMEEGDDAED SQGESEEDRA GDRNSEDDGV VMTFSSVKVS EEVEKGRAVK NQIALWDQLL EG RIKLQKA LLTTNQLPQP DVFPLFKDKG GPEFSSALKN SHKALKALLR SLVGLQEELL FQYPDTRYLV DGTKPNAGSE EIS SEDDEL VEEKKQQRRR VPAKRKLEME DYPSFMAKRF ADFTVYRNRT LQKWHDKTKL ASGKLGKGFG AFERSILTQI DHIL MDKER LLRRTQTKRS VYRVLGKPEP AAQPVPESLP GEPEILPQAP ANAHLKDLDE EIFDDDDFYH QLLRELIERK TSSLD PNDQ VAMGRQWLAI QKLRSKIHKK VDRKASKGRK LRFHVLSKLL SFMAPIDHTT MNDDARTELY RSLFGQLHPP DEGHGD UniProtKB: Protein AATF |
+Macromolecule #38: 40S ribosomal protein S15a
| Macromolecule | Name: 40S ribosomal protein S15a / type: protein_or_peptide / ID: 38 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 14.865555 KDa |
| Sequence | String: MVRMNVLADA LKSINNAEKR GKRQVLIRPC SKVIVRFLTV MMKHGYIGEF EIIDDHRAGK IVVNLTGRLN KCGVISPRFD VQLKDLEKW QNNLLPSRQF GFIVLTTSAG IMDHEEARRK HTGGKILGFF F UniProtKB: Small ribosomal subunit protein uS8 |
+Macromolecule #39: 40S ribosomal protein S27
| Macromolecule | Name: 40S ribosomal protein S27 / type: protein_or_peptide / ID: 39 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 9.480186 KDa |
| Sequence | String: MPLAKDLLHP SPEEEKRKHK KKRLVQSPNS YFMDVKCPGC YKITTVFSHA QTVVLCVGCS TVLCQPTGGK ARLTEGCSFR RKQH UniProtKB: Small ribosomal subunit protein eS27 |
+Macromolecule #40: RRP12-like protein
| Macromolecule | Name: RRP12-like protein / type: protein_or_peptide / ID: 40 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 73.292844 KDa |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) ...String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) |
+Macromolecule #41: Ubiquitin-40S ribosomal protein S27a
| Macromolecule | Name: Ubiquitin-40S ribosomal protein S27a / type: protein_or_peptide / ID: 41 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 18.004041 KDa |
| Sequence | String: MQIFVKTLTG KTITLEVEPS DTIENVKAKI QDKEGIPPDQ QRLIFAGKQL EDGRTLSDYN IQKESTLHLV LRLRGGAKKR KKKSYTTPK KNKHKRKKVK LAVLKYYKVD ENGKISRLRR ECPSDECGAG VFMASHFDRH YCGKCCLTYC FNKPEDK UniProtKB: Ubiquitin-ribosomal protein eS31 fusion protein |
+Macromolecule #42: 40S ribosomal protein S17
| Macromolecule | Name: 40S ribosomal protein S17 / type: protein_or_peptide / ID: 42 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 15.578156 KDa |
| Sequence | String: MGRVRTKTVK KAARVIIEKY YTRLGNDFHT NKRVCEEIAI IPSKKLRNKI AGYVTHLMKR IQRGPVRGIS IKLQEEERER RDNYVPEVS ALDQEIIEVD PDTKEMLKLL DFGSLSNLQV TQPTVGMNFK TPRGPV UniProtKB: Small ribosomal subunit protein eS17 |
+Macromolecule #43: Nucleolar protein 10
| Macromolecule | Name: Nucleolar protein 10 / type: protein_or_peptide / ID: 43 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 80.431633 KDa |
| Sequence | String: MQVSSLNEVK IYSLSCGKSL PEWLSDRKKR ALQKKDVDVR RRIELIQDFE MPTVCTTIKV SKDGQYILAT GTYKPRVRCY DTYQLSLKF ERCLDSEVVT FEILSDDYSK IVFLHNDRYI EFHSQSGFYY KTRIPKFGRD FSYHYPSCDL YFVGASSEVY R LNLEQGRY ...String: MQVSSLNEVK IYSLSCGKSL PEWLSDRKKR ALQKKDVDVR RRIELIQDFE MPTVCTTIKV SKDGQYILAT GTYKPRVRCY DTYQLSLKF ERCLDSEVVT FEILSDDYSK IVFLHNDRYI EFHSQSGFYY KTRIPKFGRD FSYHYPSCDL YFVGASSEVY R LNLEQGRY LNPLQTDAAE NNVCDINSVH GLFATGTIEG RVECWDPRTR NRVGLLDCAL NSVTADSEIN SLPTISALKF NG ALTMAVG TTTGQVLLYD LRSDKPLLVK DHQYGLPIKS VHFQDSLDLI LSADSRIVKM WNKNSGKIFT SLEPEHDLND VCL YPNSGM LLTANETPKM GIYYIPVLGP APRWCSFLDN LTEELEENPE STVYDDYKFV TKKDLENLGL THLIGSPFLR AYMH GFFMD IRLYHKVKLM VNPFAYEEYR KDKIRQKIEE TRAQRVQLKK LPKVNKELAL KLIEEEEEKQ KSTWKKKVKS LPNIL TDDR FKVMFENPDF QVDEESEEFR LLNPLVSKIS EKRKKKLRLL EQQELREKEE EEEPEGKPSD AESSESSDDE KAWVEE VRK QRRLLQQEEK VKRQERLKED QQTVLKPQFY EIKAGEEFRS FKDSATKQKL MNKTLEDRLK IEAKNGTLSV SDTTVGS KQ LTFTLKRSEQ QKKQQEAEKL HRQERKRLRR SAGHLKSRHK RGRSFH UniProtKB: Nucleolar protein 10 |
+Macromolecule #44: Nucleolar protein 56
| Macromolecule | Name: Nucleolar protein 56 / type: protein_or_peptide / ID: 44 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 66.160062 KDa |
| Sequence | String: MVLLHVLFEH AVGYALLALK EVEEISLLQP QVEESVLNLG KFHSIVRLVA FCPFASSQVA LENANAVSEG VVHEDLRLLL ETHLPSKKK KVLLGVGDPK IGAAIQEELG YNCQTGGVIA EILRGVRLHF HNLVKGLTDL SACKAQLGLG HSYSRAKVKF N VNRVDNMI ...String: MVLLHVLFEH AVGYALLALK EVEEISLLQP QVEESVLNLG KFHSIVRLVA FCPFASSQVA LENANAVSEG VVHEDLRLLL ETHLPSKKK KVLLGVGDPK IGAAIQEELG YNCQTGGVIA EILRGVRLHF HNLVKGLTDL SACKAQLGLG HSYSRAKVKF N VNRVDNMI IQSISLLDQL DKDINTFSMR VREWYGYHFP ELVKIINDNA TYCRLAQFIG NRRELNEDKL EKLEELTMDG AK AKAILDA SRSSMGMDIS AIDLINIESF SSRVVSLSEY RQSLHTYLRS KMSQVAPSLS ALIGEAVGAR LIAHAGSLTN LAK YPASTV QILGAEKALF RALKTRGNTP KYGLIFHSTF IGRAAAKNKG RISRYLANKC SIASRIDCFS EVPTSVFGEK LREQ VEERL SFYETGEIPR KNLDVMKEAM VQAEEAAAEI TRKLEKQEKK RLKKEKKRLA ALALASSENS SSTPEECEEM SEKPK KKKK QKPQEVPQEN GMEDPSISFS KPKKKKSFSK EELMSSDLEE TAGSTSIPKR KKSTPKEETV NDPEEAGHRS GSKKKR KFS KEEPVSSGPE EAVGKSSSKK KKKFHKASQE D UniProtKB: Nucleolar protein 56 |
+Macromolecule #45: Nucleolar protein 58
| Macromolecule | Name: Nucleolar protein 58 / type: protein_or_peptide / ID: 45 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 59.686332 KDa |
| Sequence | String: MLVLFETSVG YAIFKVLNEK KLQEVDSLWK EFETPEKANK IVKLKHFEKF QDTAEALAAF TALMEGKINK QLKKVLKKIV KEAHEPLAV ADAKLGGVIK EKLNLSCIHS PVVNELMRGI RSQMDGLIPG VEPREMAAMC LGLAHSLSRY RLKFSADKVD T MIVQAISL ...String: MLVLFETSVG YAIFKVLNEK KLQEVDSLWK EFETPEKANK IVKLKHFEKF QDTAEALAAF TALMEGKINK QLKKVLKKIV KEAHEPLAV ADAKLGGVIK EKLNLSCIHS PVVNELMRGI RSQMDGLIPG VEPREMAAMC LGLAHSLSRY RLKFSADKVD T MIVQAISL LDDLDKELNN YIMRCREWYG WHFPELGKII SDNLTYCKCL QKVGDRKNYA SAKLSELLPE EVEAEVKAAA EI SMGTEVS EEDICNILHL CTQVIEISEY RTQLYEYLQN RMMAIAPNVT VMVGELVGAR LIAHAGSLLN LAKHAASTVQ ILG AEKALF RALKSRRDTP KYGLIYHASL VGQTSPKHKG KISRMLAAKT VLAIRYDAFG EDSSSAMGVE NRAKLEARLR TLED RGIRK ISGTGKALAK TEKYEHKSEV KTYDPSGDST LPTCSKKRKI EQVDKEDEIT EKKAKKAKIK VKVEEEEEEK VAEEE ETSV KKKKKRGKKK HIKEEPLSEE EPCTSTAIAS PEKKKKKKKK RENED UniProtKB: Nucleolar protein 58 |
+Macromolecule #46: rRNA 2'-O-methyltransferase fibrillarin
| Macromolecule | Name: rRNA 2'-O-methyltransferase fibrillarin / type: protein_or_peptide / ID: 46 / Number of copies: 2 / Enantiomer: LEVO EC number: Transferases; Transferring one-carbon groups; Methyltransferases |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 33.84334 KDa |
| Sequence | String: MKPGFSPRGG GFGGRGGFGD RGGRGGRGGF GGGRGRGGGF RGRGRGGGGG GGGGGGGGRG GGGFHSGGNR GRGRGGKRGN QSGKNVMVE PHRHEGVFIC RGKEDALVTK NLVPGESVYG EKRVSISEGD DKIEYRAWNP FRSKLAAAIL GGVDQIHIKP G AKVLYLGA ...String: MKPGFSPRGG GFGGRGGFGD RGGRGGRGGF GGGRGRGGGF RGRGRGGGGG GGGGGGGGRG GGGFHSGGNR GRGRGGKRGN QSGKNVMVE PHRHEGVFIC RGKEDALVTK NLVPGESVYG EKRVSISEGD DKIEYRAWNP FRSKLAAAIL GGVDQIHIKP G AKVLYLGA ASGTTVSHVS DIVGPDGLVY AVEFSHRSGR DLINLAKKRT NIIPVIEDAR HPHKYRMLIA MVDVIFADVA QP DQTRIVA LNAHTFLRNG GHFVISIKAN CIDSTASAEA VFASEVKKMQ QENMKPQEQL TLEPYERDHA VVVGVYRPPP KVK N UniProtKB: rRNA 2'-O-methyltransferase fibrillarin |
+Macromolecule #47: NHP2-like protein 1
| Macromolecule | Name: NHP2-like protein 1 / type: protein_or_peptide / ID: 47 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 14.191524 KDa |
| Sequence | String: MTEADVNPKA YPLADAHLTK KLLDLVQQSC NYKQLRKGAN EATKTLNRGI SEFIVMAADA EPLEIILHLP LLCEDKNVPY VFVRSKQAL GRACGVSRPV IACSVTIKEG SQLKQQIQSI QQSIERLLV UniProtKB: NHP2-like protein 1 |
+Macromolecule #48: RNA 3'-terminal phosphate cyclase-like protein
| Macromolecule | Name: RNA 3'-terminal phosphate cyclase-like protein / type: protein_or_peptide / ID: 48 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.893574 KDa |
| Sequence | String: MATQAHSLSY AGCNFLRQRL VLSTLSGRPV KIRKIRARDD NPGLRDFEAS FIRLLDKITN GSRIEINQTG TTLYYQPGLL YGGSVEHDC SVLRGIGYYL ESLLCLAPFM KHPLKIVLRG VTNDQVDPSV DVLKATALPL LKQFGIDGES FELKIVRRGM P PGGGGEVV ...String: MATQAHSLSY AGCNFLRQRL VLSTLSGRPV KIRKIRARDD NPGLRDFEAS FIRLLDKITN GSRIEINQTG TTLYYQPGLL YGGSVEHDC SVLRGIGYYL ESLLCLAPFM KHPLKIVLRG VTNDQVDPSV DVLKATALPL LKQFGIDGES FELKIVRRGM P PGGGGEVV FSCPVRKVLK PIQLTDPGKI KRIRGMAYSV RVSPQMANRI VDSARSILNK FIPDIYIYTD HMKGVNSGKS PG FGLSLVA ETTSGTFLSA ELASNPQGQG AAVLPEDLGR NCARLLLEEI YRGGCVDSTN QSLALLLMTL GQQDVSKVLL GPL SPYTIE FLRHLKSFFQ IMFKIETKPC GEELKGGDKV LMTCVGIGFS NLSKTLK UniProtKB: RNA 3'-terminal phosphate cyclase-like protein |
+Macromolecule #49: Ribosome biogenesis protein BMS1 homolog
| Macromolecule | Name: Ribosome biogenesis protein BMS1 homolog / type: protein_or_peptide / ID: 49 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 146.044984 KDa |
| Sequence | String: MEAKDQKKHR KKNSGPKAAK KKKRLLQDLQ LGDEEDARKR NPKAFAVQSA VRMARSFHRT QDLKTKKHHI PVVDRTPLEP PPIVVVVMG PPKVGKSTLI QCLIRNFTRQ KLTEIRGPVT IVSGKKRRLT IIECGCDINM MIDLAKVADL VLMLIDASFG F EMETFEFL ...String: MEAKDQKKHR KKNSGPKAAK KKKRLLQDLQ LGDEEDARKR NPKAFAVQSA VRMARSFHRT QDLKTKKHHI PVVDRTPLEP PPIVVVVMG PPKVGKSTLI QCLIRNFTRQ KLTEIRGPVT IVSGKKRRLT IIECGCDINM MIDLAKVADL VLMLIDASFG F EMETFEFL NICQVHGFPK IMGVLTHLDS FKHNKQLKKT KKRLKHRFWT EVYPGAKLFY LSGMVHGEYQ NQEIHNLGRF IT VMKFRPL TWQTSHPYIL ADRMEDLTNP EDIRTNIKCD RKVSLYGYLR GAHLKNKSQI HMPGVGDFAV SDISFLPDPC ALP EQQKKR CLNEKEKLVY APLSGVGGVL YDKDAVYVDL GGSHVFQDEV GPTHELVQSL ISTHSTIDAK MASSRVTLFS DSKP LGSED IDNQGLMMPK EEKQMDLNTG RMRRKAIFGD EDESGDSDDE EDDEMSEDDG LENGSSDEEA EEEENAEMTD QYMAV KGIK RRKLELEEDS EMDLPAFADS DDDLERSSAE EGEAEEADES SEEEDCTAGE KGISGSKAAG EGSKAGLSPA NCQSDR VNL EKSLLMKKAA LPTFDSGHCT AEEVFASEDE SEESSSLSAE EEDSENEEAI RKKLSKPSQV SSGQKLGPQN FIDETSD IE NLLKEEEDYK EENNDSKETS GALKWKEDLS RKAAEAFLRQ QQAAPNLRKL IYGTVTEDNE EEDDDTLEEL GGLFRVNQ P DRECKHKADS LDCSRFLVEA PHDWDLEEVM NSIRDCFVTG KWEDDKDAAK VLAEDEELYG DFEDLETGDV HKGKSGPNT QNEDIEKEVK EEIDPDEEES AKKKHLDKKR KLKEMFDAEY DEGESTYFDD LKGEMQKQAQ LNRAEFEDQD DEARVQYEGF RPGMYVRIE IENVPCEFVQ NFDPHYPIIL GGLGNSEGNV GYVQMRLKKH RWYKKILKSR DPIIFSVGWR RFQTIPLYYI E DHNGRQRL LKYTPQHMHC GAAFWGPITP QGTGFLAIQS VSGIMPDFRI AATGVVLDLD KSIKIVKKLK LTGFPYKIFK NT SFIKGMF NSALEVAKFE GAVIRTVSGI RGQIKKALRA PEGAFRASFE DKLLMSDIVF MRTWYPVSIP AFYNPVTSLL KPV GEKDTW SGMRTTGQLR LAHGVRLKAN KDSLYKPILR QKKHFNSLHI PKALQKALPF KNKPKTQAKA GKVPKDRRRP AVIR EPHER KILALLDALS TVHSQKMKKA KEQRHLHNKE HFRAKQKEEE EKLKRQKDLR KKLFRIQGQK ERRNQKSSLK GAEGQ LQ UniProtKB: Ribosome biogenesis protein BMS1 homolog |
+Macromolecule #50: Ribosomal RNA small subunit methyltransferase NEP1
| Macromolecule | Name: Ribosomal RNA small subunit methyltransferase NEP1 / type: protein_or_peptide / ID: 50 / Number of copies: 2 / Enantiomer: LEVO EC number: Transferases; Transferring one-carbon groups; Methyltransferases |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 26.757143 KDa |
| Sequence | String: MAAPSDGFKP RERSGGEQAQ DWDALPPKRP RLGAGNKIGG RRLIVVLEGA SLETVKVGKT YELLNCDKHK SILLKNGRDP GEARPDITH QSLLMLMDSP LNRAGLLQVY IHTQKNVLIE VNPQTRIPRT FDRFCGLMVQ LLHKLSVRAA DGPQKLLKVI K NPVSDHFP ...String: MAAPSDGFKP RERSGGEQAQ DWDALPPKRP RLGAGNKIGG RRLIVVLEGA SLETVKVGKT YELLNCDKHK SILLKNGRDP GEARPDITH QSLLMLMDSP LNRAGLLQVY IHTQKNVLIE VNPQTRIPRT FDRFCGLMVQ LLHKLSVRAA DGPQKLLKVI K NPVSDHFP VGCMKVGTSF SIPVVSDVRE LVPSSDPIVF VVGAFAHGKV SVEYTEKMVS ISNYPLSAAL TCAKLTTAFE EV WGVI UniProtKB: Ribosomal RNA small subunit methyltransferase NEP1 |
+Macromolecule #51: rRNA-processing protein FCF1 homolog
| Macromolecule | Name: rRNA-processing protein FCF1 homolog / type: protein_or_peptide / ID: 51 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.416748 KDa |
| Sequence | String: MGKQKKTRKY ATMKRMLSLR DQRLKEKDRL KPKKKEKKDP SALKEREVPQ HPSCLFFQYN TQLGPPYHIL VDTNFINFSI KAKLDLVQS MMDCLYAKCI PCITDCVMAE IEKLGQKYRV ALRIAKDPRF ERLPCTHKGT YADDCLVQRV TQHKCYIVAT V DRDLKRRI ...String: MGKQKKTRKY ATMKRMLSLR DQRLKEKDRL KPKKKEKKDP SALKEREVPQ HPSCLFFQYN TQLGPPYHIL VDTNFINFSI KAKLDLVQS MMDCLYAKCI PCITDCVMAE IEKLGQKYRV ALRIAKDPRF ERLPCTHKGT YADDCLVQRV TQHKCYIVAT V DRDLKRRI RKIPGVPIMY ISNHRYNIER MPDDYGAPRF UniProtKB: rRNA-processing protein FCF1 homolog |
+Macromolecule #52: U3 small nucleolar ribonucleoprotein protein IMP4
| Macromolecule | Name: U3 small nucleolar ribonucleoprotein protein IMP4 / type: protein_or_peptide / ID: 52 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 33.818672 KDa |
| Sequence | String: MLRREARLRR EYLYRKAREE AQRSAQERKE RLRRALEENR LIPTELRREA LALQGSLEFD DAGGEGVTSH VDDEYRWAGV EDPKVMITT SRDPSSRLKM FAKELKLVFP GAQRMNRGRH EVGALVRACK ANGVTDLLVV HEHRGTPVGL IVSHLPFGPT A YFTLCNVV ...String: MLRREARLRR EYLYRKAREE AQRSAQERKE RLRRALEENR LIPTELRREA LALQGSLEFD DAGGEGVTSH VDDEYRWAGV EDPKVMITT SRDPSSRLKM FAKELKLVFP GAQRMNRGRH EVGALVRACK ANGVTDLLVV HEHRGTPVGL IVSHLPFGPT A YFTLCNVV MRHDIPDLGT MSEAKPHLIT HGFSSRLGKR VSDILRYLFP VPKDDSHRVI TFANQDDYIS FRHHVYKKTD HR NVELTEV GPRFELKLYM IRLGTLEQEA TADVEWRWHP YTNTARKRVF LSTE UniProtKB: U3 small nucleolar ribonucleoprotein protein IMP4 |
+Macromolecule #53: Deoxynucleotidyltransferase terminal-interacting protein 2
| Macromolecule | Name: Deoxynucleotidyltransferase terminal-interacting protein 2 type: protein_or_peptide / ID: 53 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 84.598102 KDa |
| Sequence | String: MVVTRSARAK ASIQAASAES SGQKSFAANG IQAHPESSTG SDARTTAESQ TTGKQSLIPR TPKARKRKSR TTGSLPKGTE PSTDGETSE AESNYSVSEH HDTILRVTRR RQILIACSPV SSVRKKPKVT PTKESYTEEI VSEAESHVSG ISRIVLPTEK T TGARRSKA ...String: MVVTRSARAK ASIQAASAES SGQKSFAANG IQAHPESSTG SDARTTAESQ TTGKQSLIPR TPKARKRKSR TTGSLPKGTE PSTDGETSE AESNYSVSEH HDTILRVTRR RQILIACSPV SSVRKKPKVT PTKESYTEEI VSEAESHVSG ISRIVLPTEK T TGARRSKA KSLTDPSQES HTEAISDAET SSSDISFSGI ATRRTRSMQR KLKAQTEKKD SKIVPGNEKQ IVGTPVNSED SD TRQTSHL QARSLSEINK PNFYNNDFDD DFSHRSSENI LTVHEQANVE SLKETKQNCK DLDEDANGIT DEGKEINEKS SQL KNLSEL QDTSLQQLVS QRHSTPQNKN AVSVHSNLNS EAVMKSLTQT FATVEVGRWN NNKKSPIKAS DLTKFGDCGG SDDE EESTV ISVSEDMNSE GNVDFECDTK LYTSAPNTSQ GKDNSVLLVL SSDESQQSEN SENEEDTLCF VENSGQRESL SGDTG SLSC DNALFVIDTT PGMSADKNFY LEEEDKASEV AIEEEKEEEE DEKSEEDSSD HDENEDEFSD EEDFLNSTKA KLLKLT SSS IDPGLSIKQL GGLYINFNAD KLQSNKRTLT QIKEKKKNEL LQKAVITPDF EKNHCVPPYS ESKYQLQKKR RKERQKT AG DGWFGMKAPE MTNELKNDLK ALKMRASMDP KRFYKKNDRD GFPKYFQIGT IVDNPADFYH SRIPKKQRKR TIVEELLA D SEFRRYNRRK YSEIMAEKAA NAAGKKFRKK KKFRN UniProtKB: Deoxynucleotidyltransferase terminal-interacting protein 2 |
+Macromolecule #54: 40S ribosomal protein S23
| Macromolecule | Name: 40S ribosomal protein S23 / type: protein_or_peptide / ID: 54 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 15.844666 KDa |
| Sequence | String: MGKCRGLRTA RKLRSHRRDQ KWHDKQYKKA HLGTALKANP FGGASHAKGI VLEKVGVEAK QPNSAIRKCV RVQLIKNGKK ITAFVPNDG CLNFIEENDE VLVAGFGRKG HAVGDIPGVR FKVVKVANVS LLALYKGKKE RPRS UniProtKB: Small ribosomal subunit protein uS12 |
+Macromolecule #55: U3 small nucleolar RNA-associated protein 14 homolog A
| Macromolecule | Name: U3 small nucleolar RNA-associated protein 14 homolog A type: protein_or_peptide / ID: 55 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 88.129125 KDa |
| Sequence | String: MTANRLAESL LALSQQEELA DLPKDYLLSE SEDEGDNDGE RKHQKLLEAI SSLDGKNRRK LAERSEASLK VSEFNVSSEG SGEKLVLAD LLEPVKTSSS LATVKKQLSR VKSKKTVELP LNKEEIERIH REVAFNKTAQ VLSKWDPVVL KNRQAEQLVF P LEKEEPAI ...String: MTANRLAESL LALSQQEELA DLPKDYLLSE SEDEGDNDGE RKHQKLLEAI SSLDGKNRRK LAERSEASLK VSEFNVSSEG SGEKLVLAD LLEPVKTSSS LATVKKQLSR VKSKKTVELP LNKEEIERIH REVAFNKTAQ VLSKWDPVVL KNRQAEQLVF P LEKEEPAI APIEHVLSGW KARTPLEQEI FNLLHKNKQP VTDPLLTPVE KASLRAMSLE EAKMRRAELQ RARALQSYYE AK ARREKKI KSKKYHKVVK KGKAKKALKE FEQLRKVNPA AALEELEKIE KARMMERMSL KHQNSGKWAK SKAIMAKYDL EAR QAMQEQ LSKNKELTQK LQVASESEEE EGGTEDVEEL LVPDVVNEVQ MNADGPNPWM LRSCTSDTKE AATQEDPEQL PELE AHGVS ESEGEERPVA EEEILLREFE ERRSLRKRSE LSQDAEPAGS QETKDSGSQE VLSELRVLSQ KLKENHQSRK QKASS EGTI PQVQREEPAP EEEEPLLLQR PERVQTLEEL EELGKEECFQ NKELPRPVLE GQQSERTPNN RPDAPKEKKK KEQMID LQN LLTTQSPSVK SLAVPTIEEL EDEEERNHRQ MIKEAFAGDD VIRDFLKEKR EAVEASKPKD VDLTLPGWGE WGGVGLK PS AKKRRRFLIK APEGPPRKDK NLPNVIINEK RNIHAAAHQV RVLPYPFTHH WQFERTIQTP IGSTWNTQRA FQKLTTPK V VTKPGHIINP IKAEDVGYRS SSRSDLSVIQ RNPKRITTRH KKQLKKCSVD UniProtKB: U3 small nucleolar RNA-associated protein 14 homolog A |
+Macromolecule #56: Unassigned peptides
| Macromolecule | Name: Unassigned peptides / type: protein_or_peptide / ID: 56 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 15.081565 KDa |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) ...String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) |
+Macromolecule #57: Probable U3 small nucleolar RNA-associated protein 11
| Macromolecule | Name: Probable U3 small nucleolar RNA-associated protein 11 / type: protein_or_peptide / ID: 57 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 30.520779 KDa |
| Sequence | String: MAAAFRKAAK SRQREHRERS QPGFRKHLGL LEKKKDYKLR ADDYRKKQEY LKALRKKALE KNPDEFYYKM TRVKLQDGVH IIKETKEEV TPEQLKLMRT QDVKYIEMKR VAEAKKIERL KSELHLLDFQ GKQQNKHVFF FDTKKEVEQF DVATHLQTAP E LVDRVFNR ...String: MAAAFRKAAK SRQREHRERS QPGFRKHLGL LEKKKDYKLR ADDYRKKQEY LKALRKKALE KNPDEFYYKM TRVKLQDGVH IIKETKEEV TPEQLKLMRT QDVKYIEMKR VAEAKKIERL KSELHLLDFQ GKQQNKHVFF FDTKKEVEQF DVATHLQTAP E LVDRVFNR PRIETLQKEK VKGVTNQTGL KRIAKERQKQ YNCLTQRIER EKKLFVIAQK IQTRKDLMDK TQKVKVKKET VN SPAIYKF QSRRKR UniProtKB: Probable U3 small nucleolar RNA-associated protein 11 |
+Macromolecule #58: Nucleolar protein 6
| Macromolecule | Name: Nucleolar protein 6 / type: protein_or_peptide / ID: 58 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 127.748641 KDa |
| Sequence | String: MGPAPAGEQL RGATGEPEVM EPALEGTGKE GKKASSRKRT LAEPPAKGLL QPVKLSRAEL YKEPTNEELN RLRETEILFH SSLLRLQVE ELLKEVRLSE KKKDRIDAFL REVNQRVVRV PSVPETELTD QAWLPAGVRV PLHQVPYAVK GCFRFLPPAQ V TVVGSYLL ...String: MGPAPAGEQL RGATGEPEVM EPALEGTGKE GKKASSRKRT LAEPPAKGLL QPVKLSRAEL YKEPTNEELN RLRETEILFH SSLLRLQVE ELLKEVRLSE KKKDRIDAFL REVNQRVVRV PSVPETELTD QAWLPAGVRV PLHQVPYAVK GCFRFLPPAQ V TVVGSYLL GTCIRPDINV DVALTMPREI LQDKDGLNQR YFRKRALYLA HLAHHLAQDP LFGSVCFSYT NGCHLKPSLL LR PRGKDER LVTVRLHPCP PPDFFRPCRL LPTKNNVRSA WYRGQSPAGD GSPEPPTPRY NTWVLQDTVL ESHLQLLSTI LSS AQGLKD GVALLKVWLR QRELDKGQGG FTGFLVSMLV VFLVSTRKIH TTMSGYQVLR SVLQFLATTD LTVNGISLCL SSDP SLPAL ADFHQAFSVV FLDSSGHLNL CADVTASTYH QVQHEARLSM MLLDSRADDG FHLLLMTPKP MIRAFDHVLH LRPLS RLQA ACHRLKLWPE LQDNGGDYVS AALGPLTTLL EQGLGARLNL LAHSRPPVPE WDISQDPPKH KDSGTLTLGL LLRPEG LTS VLELGPEADQ PEAAKFRQFW GSRSELRRFQ DGAIREAVVW EAASMSQKRL IPHQVVTHLL ALHADIPETC VHYVGGP LD ALIQGLKETS STGEEALVAA VRCYDDLSRL LWGLEGLPLT VSAVQGAHPV LRYTEVFPPT PVRPAFSFYE TLRERSSL L PRLDKPCPAY VEPMTVVCHL EGSGQWPQDA EAVQRVRAAF QLRLAELLTQ QHGLQCRATA THTDVLKDGF VFRIRVAYQ REPQILKEVQ SPEGMISLRD TAASLRLERD TRQLPLLTSA LHGLQQQHPA FSGVARLAKR WVRAQLLGEG FADESLDLVA AALFLHPEP FTPPSSPQVG FLRFLFLVST FDWKNNPLFV NLNNELTVEE QVEIRSGFLA ARAQLPVMVI VTPQDRKNSV W TQDGPSAQ ILQQLVVLAA EALPMLEKQL MDPRGPGDIR TVFRPPLDIY DVLIRLSPRH IPRHRQAVDS PAASFCRGLL SQ PGPSSLM PVLGYDPPQL YLTQLREAFG DLALFFYDQH GGEVIGVLWK PTSFQPQPFK ASSTKGRMVM SRGGELVMVP NVE AILEDF AVLGEGLVQT VEARSERWTV UniProtKB: Nucleolar protein 6 |
+Macromolecule #59: Small subunit processome component 20 homolog
| Macromolecule | Name: Small subunit processome component 20 homolog / type: protein_or_peptide / ID: 59 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 318.81325 KDa |
| Sequence | String: MKTKPVSHKT ENTYRFLTFA ERLGNVNIDI IHRIDRTASY EEEVETYFFE GLLKWRELNL TEHFGKFYKE VIDKCQSFNQ LVYHQNEIV QSLKTHLQVK NSFAYQPLLD LVVQLARDLQ MDFYPHFPEF FLTITSILET QDTELLEWAF TSLSYLYKYL W RLMVKDMS ...String: MKTKPVSHKT ENTYRFLTFA ERLGNVNIDI IHRIDRTASY EEEVETYFFE GLLKWRELNL TEHFGKFYKE VIDKCQSFNQ LVYHQNEIV QSLKTHLQVK NSFAYQPLLD LVVQLARDLQ MDFYPHFPEF FLTITSILET QDTELLEWAF TSLSYLYKYL W RLMVKDMS SIYSMYSTLL AHKKLHIRNF AAESFTFLMR KVSDKNALFN LMFLDLDKHP EKVEGVGQLL FEMCKGVRNM FH SCTGQAV KLILRKLGPV TETETQLPWM LIGETLKNMV KSTVSYISKE HFGTFFECLQ ESLLDLHTKV TKTNCCESSE QIK RLLETY LILVKHGSGT KIPTPADVCK VLSQTLQVAS LSTSCWETLL DVISALILGE NVSLPETLIK ETIEKIFESR FEKR LIFSF SEVMFAMKQF EQLFLPSFLS YIVNCFLIDD AVVKDEALAI LAKLILNKAA PPTAGSMAIE KYPLVFSPQM VGFYI KQKK TRSKGRNEQF PVLDHLLSII KLPPNKDTTY LSQSWAALVV LPHIRPLEKE KVIPLVTGFI EALFMTVDKG SFGKGN LFV LCQAVNTLLS LEESSELLHL VPVERVKNLV LTFPLEPSVL LLTDLYYQRL ALCGCKGPLS QEALMELFPK LQANIST GV SKIRLLTIRI LNHFDVQLPE SMEDDGLSER QSVFAILRQA ELVPATVNDY REKLLHLRKL RHDVVQTAVP DGPLQEVP L RYLLGMLYIN FSALWDPVIE LISSHAHEME NKQFWKVYYE HLEKAATHAE KELQNDMTDE KSVGDESWEQ TQEGDVGAL YHEQLALKTD CQERLDHTNF RFLLWRALTK FPERVEPRSR ELSPLFLRFI NNEYYPADLQ VAPTQDLRRK GKGMVAEEIE EEPAAGDDE ELEEEAVPQD ESSQKKKTRR AAAKQLIAHL QVFSKFSNPR ALYLESKLYE LYLQLLLHQD QMVQKITLDC I MTYKHPHV LPYRENLQRL LEDRSFKEEI VHFSISEDNA VVKTAHRADL FPILMRILYG RMKNKTGSKT QGKSASGTRM AI VLRFLAG TQPEEIQIFL DLLFEPVRHF KNGECHSAVI QAVEDLDLSK VLPLGRQHGI LNSLEIVLKN ISHLISAYLP KIL QILLCM TATVSHILDQ REKIQLRFIN PLKNLRRLGI KMVTDIFLDW ESYQFRTEEI DAVFHGAVWP QISRLGSESQ YSPT PLLKL ISIWSRNARY FPLLAKQKPG HPECDILTNV FAILSAKNLS DATASIVMDI VDDLLNLPDF EPTETVLNLL VTGCV YPGI AENIGESITI GGRLILPHVP AILQYLSKTT ISAEKVKKKK NRAQVSKELG ILSKISKFMK DKEQSSVLIT LLLPFL HRG NIAEDTEVDI LVTVQNLLKH CVDPTSFLKP IAKLFSVIKN KLSRKLLCTV FETLSDFESG LKYITDVVKL NAFDQRH LD DINFDVRFET FQTITSYIKE MQIVDVNYLI PVMHNCFYNL ELGDMSLSDN ASMCLMSIIK KLAALNVTEK DYREIIHR S LLEKLRKGLK SQTESIQQDY TTILSCLIQT FPNQLEFKDL VQLTHYHDPE MDFFENMKHI QIHRRARALK KLAKQLMEG KVVLSSKSLQ NYIMPYAMTP IFDEKMLKHE NITTAATEII GAICKHLSWS AYMYYLKHFI HVLQTGQINQ KLGVSLLVIV LEAFHFDHK TLEEQMGKIE NEENAIEAIE LPEPEAMELE RVDEEEKEYT CKSLSDNGQP GTPDPADSGG TSAKESECIT K PVSFLPQN KEEIERTIKN IQGTITGDIL PRLHKCLAST TKREEEHKLV KSKVVNDEEV VRVPLAFAMV KLMQSLPQEV ME ANLPSIL LKVCALLKNR AQEIRDIARS TLAKIIEDLG VHFLLYVLKE LQTTLVRGYQ VHVLTFTVHM LLQGLTNKLQ VGD LDSCLD IMIEIFNHEL FGAVAEEKEV KQILSKVMEA RRSKSYDSYE ILGKFVGKDQ VTKLILPLKE ILQNTTSLKL ARKV HETLR RITVGLIVNQ EMTAESILLL SYGLISENLP LLTEKEKNPV APAPDPRLPP QSCLLLPPTP VRGGQKAVVS RKTNM HIFI ESGLRLLHLS LKTSKIKSSG ECVLEMLDPF VSLLIDCLGS MDVKVITGAL QCLIWVLRFP LPSIETKAEQ LTKHLF LLL KDYAKLGAAR GQNFHLVVNC FKCVTILVKK VKSYQITEKQ LQVLLAYAEE DIYDTSRQAT AFGLLKAILS RKLLVPE ID EVMRKVSKLA VSAQSEPARV QCRQVFLKYI LDYPLGDKLR PNLEFMLAQL NYEHETGRES TLEMIAYLFD TFPQGLLH E NCGMFFIPLC LMTINDDSAT CKKMASMTIK SLLGKISLEK KDWLFDMVTT WFGAKKRLNR QLAALICGLF VESEGVDFE KRLGTVLPVI EKEIDPENFK DIMEETEEKA ADRLLFSFLT LITKLIKECN IIQFTKPAET LSKIWSHVHS HLRHPHNWVW LTAAQIFGL LFASCQPEEL IQKWNTKKTK KHLPEPVAIK FLASDLDQKM KSISLASCHQ LHSKFLDQSL GEQVVKNLLF A AKVLYLLE LYCEDKQSKI KEDLEEQEAL EDGVACADEK AESDGEEKEE VKEELGRPAT LLWLIQKLSR IAKLEAAYSP RN PLKRTCI FKFLGAVAMD LGIDKVKPYL PMIIAPLFRE LNSTYSEQDP LLKNLSQEII ELLKKLVGLE SFSLAFASVQ KQA NEKRAL RKKRKALEFV TNPDIAAKKK MKKHKNKSEA KKRKIEFLRP GYKAKRQKSH SLKDLAMVE UniProtKB: Small subunit processome component 20 homolog |
+Macromolecule #60: Transducin beta-like protein 3
| Macromolecule | Name: Transducin beta-like protein 3 / type: protein_or_peptide / ID: 60 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 89.14357 KDa |
| Sequence | String: MAETAAGVGR FKTNYAVERK IEPFYKGGKA QLDQTGQHLF CVCGTRVNIL EVASGAVLRS LEQEDQEDIT AFDLSPDNEV LVTASRALL LAQWAWQEGS VTRLWKAIHT APVATMAFDP TSTLLATGGC DGAVRVWDIV RHYGTHHFRG SPGVVHLVAF H PDPTRLLL ...String: MAETAAGVGR FKTNYAVERK IEPFYKGGKA QLDQTGQHLF CVCGTRVNIL EVASGAVLRS LEQEDQEDIT AFDLSPDNEV LVTASRALL LAQWAWQEGS VTRLWKAIHT APVATMAFDP TSTLLATGGC DGAVRVWDIV RHYGTHHFRG SPGVVHLVAF H PDPTRLLL FSSATDAAIR VWSLQDRSCL AVLTAHYSAV TSLAFSADGH TMLSSGRDKI CIIWDLQSCQ ATRTVPVFES VE AAVLLPE EPVSQLGVKS PGLYFLTAGD QGTLRVWEAA SGQCVYTQAQ PPGPGQELTH CTLAHTAGVV LTATADHNLL LYE ARSLRL QKQFAGYSEE VLDVRFLGPE DSHVVVASNS PCLKVFELQT SACQILHGHT DIVLALDVFR KGWLFASCAK DQSV RIWRM NKAGQVMCVA QGSGHTHSVG TVCCSRLKES FLVTGSQDCT VKLWPLPKAL LSKNTAPDNG PILLQAQTTQ RCHDK DINS VAIAPNDKLL ATGSQDRTAK LWALPQCQLL GVFSGHRRGL WCVQFSPMDQ VLATASADGT IKLWALQDFS CLKTFE GHD ASVLKVAFVS RGTQLLSSGS DGLVKLWTIK NNECVRTLDA HEDKVWGLHC SRLDDHALTG ASDSRVILWK DVTEAEQ AE EQARQEEQVV RQQELDNLLH EKRYLRALGL AISLDRPHTV LTVIQAIRRD PEACEKLEAT MLRLRRDQKE ALLRFCVT W NTNSRHCHEA QAVLGVLLRR EAPEELLAYE GVRAALEALL PYTERHFQRL SRTLQAAAFL DFLWHNMKLP VPAAAPTPW ETHKGALP UniProtKB: Transducin beta-like protein 3 |
+Macromolecule #61: HEAT repeat-containing protein 1
| Macromolecule | Name: HEAT repeat-containing protein 1 / type: protein_or_peptide / ID: 61 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 242.649672 KDa |
| Sequence | String: MTSLAQQLQR LALPQSDASL LSRDEVASLL FDPKEAATID RDTAFAIGCT GLEELLGIDP SFEQFEAPLF SQLAKTLERS VQTKAVNKQ LDENISLFLI HLSPYFLLKP AQKCLEWLIH RFHIHLYNQD SLIACVLPYH ETRIFVRVIQ LLKINNSKHR W FWLLPVKQ ...String: MTSLAQQLQR LALPQSDASL LSRDEVASLL FDPKEAATID RDTAFAIGCT GLEELLGIDP SFEQFEAPLF SQLAKTLERS VQTKAVNKQ LDENISLFLI HLSPYFLLKP AQKCLEWLIH RFHIHLYNQD SLIACVLPYH ETRIFVRVIQ LLKINNSKHR W FWLLPVKQ SGVPLAKGTL ITHCYKDLGF MDFICSLVTK SVKVFAEYPG SSAQLRVLLA FYASTIVSAL VAAEDVSDNI IA KLFPYIQ KGLKSSLPDY RAATYMIICQ ISVKVTMENT FVNSLASQII KTLTKIPSLI KDGLSCLIVL LQRQKPESLG KKP FPHLCN VPDLITILHG ISETYDVSPL LHYMLPHLVV SIIHHVTGEE TEGMDGQIYK RHLEAILTKI SLKNNLDHLL ASLL FEEYI SYSSQEEMDS NKVSLLNEQF LPLIRLLESK YPRTLDVVLE EHLKEIADLK KQELFHQFVS LSTSGGKYQF LADSD TSLM LSLNHPLAPV RILAMNHLKK IMKTSKEGVD ESFIKEAVLA RLGDDNIDVV LSAISAFEIF KEHFSSEVTI SNLLNL FQR AELSKNGEWY EVLKIAADIL IKEEILSEND QLSNQVVVCL LPFMVINNDD TESAEMKIAI YLSKSGICSL HPLLRGW EE ALENVIKSTK PGKLIGVANQ KMIELLADNI NLGDPSSMLK MVEDLISVGE EESFNLKQKV TFHVILSVLV SCCSSLKE T HFPFAIRVFS LLQKKIKKLE SVITAVEIPS EWHIELMLDR GIPVELWAHY VEELNSTQRV AVEDSVFLVF SLKKFIYAL KAPKSFPKGD IWWNPEQLKE DSRDYLHLLI GLFEMMLNGA DAVHFRVLMK LFIKVHLEDV FQLFKFCSVL WTYGSSLSNP LNCSVKTVL QTQALYVGCA MLSSQKTQCK HQLASISSPV VTSLLINLGS PVKEVRRAAI QCLQALSGVA SPFYLIIDHL I SKAEEITS DAAYVIQDLA TLFEELQREK KLKSHQKLSE TLKNLLSCVY SCPSYIAKDL MKVLQGVNGE MVLSQLLPMA EQ LLEKIQK EPTAVLKDEA MVLHLTLGKY NEFSVSLLNE DPKSLDIFIK AVHTTKELYA GMPTIQITAL EKITKPFFAA ISD EKVQQK LLRMLFDLLV NCKNSHCAQT VSSVFKGISV NAEQVRIELE PPDKAKPLGT VQQKRRQKMQ QKKSQDLESV QEVG GSYWQ RVTLILELLQ HKKKLRSPQI LVPTLFNLLS RCLEPLPQEQ GNMEYTKQLI LSCLLNICQK LSPDGGKIPK DILDE EKFN VELIVQCIRL SEMPQTHHHA LLLLGTVAGI FPDKVLHNIM SIFTFMGANV MRLDDTYSFQ VINKTVKMVI PALIQS DSG DSIEVSRNVE EIVVKIISVF VDALPHVPEH RRLPILVQLV DTLGAEKFLW ILLILLFEQY VTKTVLAAAY GEKDAIL EA DTEFWFSVCC EFSVQHQIQS LMNILQYLLK LPEEKEETIP KAVSFNKSES QEEMLQVFNV ETHTSKQLRH FKFLSVSF M SQLLSSNNFL KKVVESGGPE ILKGLEERLL ETVLGYISAV AQSMERNADK LTVKFWRALL SKAYDLLDKV NALLPTETF IPVIRGLVGN PLPSVRRKAL DLLNNKLQQN ISWKKTIVTR FLKLVPDLLA IVQRKKKEGE EEQAINRQTA LYTLKLLCKN FGAENPDPF VPVLNTAVKL IAPERKEEKN VLGSALLCIA EVTSTLEALA IPQLPSLMPS LLTTMKNTSE LVSSEVYLLS A LAALQKVV ETLPHFISPY LEGILSQVIH LEKITSEMGS ASQANIRLTS LKKTLATTLA PRVLLPAIKK TYKQIEKNWK NH MGPFMSI LQEHIGVMKK EELTSHQSQL TAFFLEALDF RAQHSENDLE EVGKTENCII DCLVAMVVKL SEVTFRPLFF KLF DWAKTE DAPKDRLLTF YNLADCIAEK LKGLFTLFAG HLVKPFADTL NQVNISKTDE AFFDSENDPE KCCLLLQFIL NCLY KIFLF DTQHFISKER AEALMMPLVD QLENRLGGEE KFQERVTKHL IPCIAQFSVA MADDSLWKPL NYQILLKTRD SSPKV RFAA LITVLALAEK LKENYIVLLP ESIPFLAELM EDECEEVEHQ CQKTIQQLET VLGEPLQSYF UniProtKB: HEAT repeat-containing protein 1 |
+Macromolecule #63: U3 small nucleolar RNA-interacting protein 2
| Macromolecule | Name: U3 small nucleolar RNA-interacting protein 2 / type: protein_or_peptide / ID: 63 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 51.920664 KDa |
| Sequence | String: MSATAAARKR GKPASGAGAG AGAGKRRRKA DSAGDRGKSK GGGKMNEEIS SDSESESLAP RKPEEEEEEE LEETAQEKKL RLAKLYLEQ LRQQEEEKAE ARAFEEDQVA GRLKEDVLEQ RGRLQKLVAK EIQAPASADI RVLRGHQLSI TCLVVTPDDS A IFSAAKDC ...String: MSATAAARKR GKPASGAGAG AGAGKRRRKA DSAGDRGKSK GGGKMNEEIS SDSESESLAP RKPEEEEEEE LEETAQEKKL RLAKLYLEQ LRQQEEEKAE ARAFEEDQVA GRLKEDVLEQ RGRLQKLVAK EIQAPASADI RVLRGHQLSI TCLVVTPDDS A IFSAAKDC SIIKWSVESG RKLHVIPRAK KGAEGKPPGH SSHVLCMAIS SDGKYLASGD RSKLILIWEA QSCQHLYTFT GH RDAVSGL AFRRGTHQLY STSHDRSVKV WNVAENSYVE TLFGHQDAVA ALDALSRECC VTAGGRDGTV RVWKIPEESQ LVF YGHQGS IDCIHLINEE HMVSGADDGS VALWGLSKKR PLALQREAHG LRGEPGLEQP FWISSVAALL NTDLVATGSH SSCV RLWQC GEGFRQLDLL CDIPLVGFIN SLKFSSSGDF LVAGVGQEHR LGRWWRIKEA RNSVCIIPLR RVPVPPAAGS UniProtKB: U3 small nucleolar RNA-interacting protein 2 |
+Macromolecule #64: Ribosomal RNA-processing protein 7 homolog A
| Macromolecule | Name: Ribosomal RNA-processing protein 7 homolog A / type: protein_or_peptide / ID: 64 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 32.39315 KDa |
| Sequence | String: MVARRRKCAA RDPEDRIPSP LGYAAIPIKF SEKQQASHYL YVRAHGVRQG TKSTWPQKRT LFVLNVPPYC TEESLSRLLS TCGLVQSVE LQEKPDLAES PKESRSKFFH PKPVPGFQVA YVVFQKPSGV SAALALKGPL LVSTESHPVK SGIHKWISDY A DSVPDPEA ...String: MVARRRKCAA RDPEDRIPSP LGYAAIPIKF SEKQQASHYL YVRAHGVRQG TKSTWPQKRT LFVLNVPPYC TEESLSRLLS TCGLVQSVE LQEKPDLAES PKESRSKFFH PKPVPGFQVA YVVFQKPSGV SAALALKGPL LVSTESHPVK SGIHKWISDY A DSVPDPEA LRVEVDTFME AYDQKIAEEE AKAKEEEGVP DEEGWVKVTR RGRRPVLPRT EAASLRVLER ERRKRSRKEL LN FYAWQHR ESKMEHLAQL RKKFEEDKQR IELLRAQRKF RPY UniProtKB: Ribosomal RNA-processing protein 7 homolog A |
+Macromolecule #65: RNA-binding protein PNO1
| Macromolecule | Name: RNA-binding protein PNO1 / type: protein_or_peptide / ID: 65 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 27.970355 KDa |
| Sequence | String: MESEMETQSA RAEEGFTQVT RKGGRRAKKR QAEQLSAAGE GGDAGRMDTE EARPAKRPVF PPLCGDGLLS GKEETRKIPV PANRYTPLK ENWMKIFTPI VEHLGLQIRF NLKSRNVEIR TCKETKDVSA LTKAADFVKA FILGFQVEDA LALIRLDDLF L ESFEITDV ...String: MESEMETQSA RAEEGFTQVT RKGGRRAKKR QAEQLSAAGE GGDAGRMDTE EARPAKRPVF PPLCGDGLLS GKEETRKIPV PANRYTPLK ENWMKIFTPI VEHLGLQIRF NLKSRNVEIR TCKETKDVSA LTKAADFVKA FILGFQVEDA LALIRLDDLF L ESFEITDV KPLKGDHLSR AIGRIAGKGG KTKFTIENVT RTRIVLADVK VHILGSFQNI KMARTAICNL ILGNPPSKVY GN IRAVASR SADRF UniProtKB: RNA-binding protein PNO1 |
+Macromolecule #66: Nucleolar protein 14
| Macromolecule | Name: Nucleolar protein 14 / type: protein_or_peptide / ID: 66 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 68.259539 KDa |
| Sequence | String: MAKAKKVGAR RKASGAPAGA RGGPAKANSN PFEVKVNRQK FQILGRKTRH DVGLPGVSRA RALRKRTQTL LKEYKERDKS NVFRDK(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK) ...String: MAKAKKVGAR RKASGAPAGA RGGPAKANSN PFEVKVNRQK FQILGRKTRH DVGLPGVSRA RALRKRTQTL LKEYKERDKS NVFRDK(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)TE AEL AKEEQE HLRKLEAERL RRML(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)AEGN KAKLEKLFGF LLEYVGDLAT DDPPDLTVID KLVVHLYHLC QMFPESASDA I KFVLRDAM HEMEEMIETK GRAALPGLDV LIYLKITGLL FPTSDFWHPV VTPALVCLSQ LLTKCPILSL QDVVKGLFVC CL FLEYVAL SQRFIPELIN FLLGILYIAT PNKASQGSTL VHPFRALGKN SELLVVSARE DVATWQQSSL SLRWASRLRA PTS TEANHI RLSCLAVGLA LLKRCVLMYG SLPSFHAIMG PLQALLTDHL ADCSHPQELQ ELCQSTLTEM ESQKQLCRPL TCEK SKPVP LKLFTPRLVK VLEFGRKQGS SKEEQERKRL IHKHKREFKG AVREIRKDNQ FLARMQLSEI MERDAERKRK VKQLF NSLA TQEGEWKALK RKKFKK |
+Macromolecule #67: Nucleolar complex protein 4 homolog
| Macromolecule | Name: Nucleolar complex protein 4 homolog / type: protein_or_peptide / ID: 67 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 50.184086 KDa |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) ...String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)PRELFKLV VGGLL(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)Y DDTRYHTMQA AVDAVARVTG QHPEVPPAF WNNAFTLLSA VSLPRREPTV SSFYVKRAEL WDTWKVAHLK EHRRVFQAMW LSFLKHKLPL SLYKKVLLIV HDAILPQLAQ PTLMIDFLT RACDLGGALS LLALNGLFIL IHKHNLEYPD FYRKLYGLLD PSVFHVKYRA RFFHLADLFL SSSHLPAYLV A AFAKRLAR LALTAPPEAL LMVLPFICNL LRRHPACRVL VHRPHGPELD ADPYDPGEED PAQSRALESS LWELQALQRH YH PEVSKAA SVINQALSMP EVSIAPLLEL TAYEIFERDL KKKGPEPVPL EFIPAQGLLG RPGELCAQHF TLS |
+Macromolecule #68: KRR1 small subunit processome component homolog
| Macromolecule | Name: KRR1 small subunit processome component homolog / type: protein_or_peptide / ID: 68 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 43.751109 KDa |
| Sequence | String: MASPSLERPE KGAGKSEFRN QKPKPENQDE SELLTVPDGW KEPAFSKEDN PRGLLEESSF ATLFPKYREA YLKECWPLVQ KALNEHHVN ATLDLIEGSM TVCTTKKTFD PYIIIRARDL IKLLARSVSF EQAVRILQDD VACDIIKIGS LVRNKERFVK R RQRLIGPK ...String: MASPSLERPE KGAGKSEFRN QKPKPENQDE SELLTVPDGW KEPAFSKEDN PRGLLEESSF ATLFPKYREA YLKECWPLVQ KALNEHHVN ATLDLIEGSM TVCTTKKTFD PYIIIRARDL IKLLARSVSF EQAVRILQDD VACDIIKIGS LVRNKERFVK R RQRLIGPK GSTLKALELL TNCYIMVQGN TVSAIGPFSG LKEVRKVVLD TMKNIHPIYN IKSLMIKREL AKDSELRSQS WE RFLPQFK HKNVNKRKEP KKKTVKKEYT PFPPPQPESQ IDKELASGEY FLKANQKKRQ KMEAIKAKQA EAISKRQEER NKA FIPPKE KPIVKPKEAS TETKIDVASI KEKVKKAKNK KLGALTAEEI ALKMEADEKK KKKKK UniProtKB: KRR1 small subunit processome component homolog |
+Macromolecule #69: Bystin
| Macromolecule | Name: Bystin / type: protein_or_peptide / ID: 69 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 33.712934 KDa |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) ...String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)PRV LEVYRGVREV LS KYRSGKL PKAFKIIPAL SNWEQILYVT EPEAWTAAAM YQATRIFASN LKERMAQRFY NLVLLPRVRD DVAEYKRLNF HLY MALKKA LFKPGAWFKG ILIPLCESGT CTLREAIIVG SIITKCSIPV LHSSAAMLKI AEMEYSGANS IFLRLLLDKK YALP YRVLD ALVFHFLGFR TEKRELPVLW HQCLLTLVQR YKADLATDQK EALLELLRLQ PHPQLSPEIR RELQSAVPRD VEDVP ITVE |
+Macromolecule #70: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 70 / Number of copies: 22 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
+Macromolecule #71: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 71 / Number of copies: 3 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
+Macromolecule #72: GUANOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: GUANOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 72 / Number of copies: 1 / Formula: GTP |
|---|---|
| Molecular weight | Theoretical: 523.18 Da |
| Chemical component information |  ChemComp-GTP: |
+Macromolecule #73: S-ADENOSYL-L-HOMOCYSTEINE
| Macromolecule | Name: S-ADENOSYL-L-HOMOCYSTEINE / type: ligand / ID: 73 / Number of copies: 2 / Formula: SAH |
|---|---|
| Molecular weight | Theoretical: 384.411 Da |
| Chemical component information |  ChemComp-SAH: |
+Macromolecule #74: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 74 / Number of copies: 2 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.6 |
|---|---|
| Grid | Model: Quantifoil R2/2 / Material: GOLD / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 3 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number real images: 84904 / Average electron dose: 58.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 0.01 mm / Nominal defocus max: 2.7 µm / Nominal defocus min: 0.7000000000000001 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL | ||||||||||||||||||||
| Output model |  PDB-7mq9: |
+ About Yorodumi
About Yorodumi
-News
-Feb 9, 2022. New format data for meta-information of EMDB entries
New format data for meta-information of EMDB entries
- Version 3 of the EMDB header file is now the official format.
- The previous official version 1.9 will be removed from the archive.
Related info.: EMDB header
EMDB header
External links: wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
-Aug 12, 2020. Covid-19 info
Covid-19 info

URL: https://pdbj.org/emnavi/covid19.php
New page: Covid-19 featured information page in EM Navigator.
Related info.: Covid-19 info /
Covid-19 info /  Mar 5, 2020. Novel coronavirus structure data
Mar 5, 2020. Novel coronavirus structure data
+Mar 5, 2020. Novel coronavirus structure data
Novel coronavirus structure data
- International Committee on Taxonomy of Viruses (ICTV) defined the short name of the 2019 coronavirus as "SARS-CoV-2".
- In the structure databanks used in Yorodumi, some data are registered as the other names, "COVID-19 virus" and "2019-nCoV". Here are the details of the virus and the list of structure data.
Related info.: Yorodumi Speices /
Yorodumi Speices /  Aug 12, 2020. Covid-19 info
Aug 12, 2020. Covid-19 info
External links: COVID-19 featured content - PDBj /
COVID-19 featured content - PDBj /  Molecule of the Month (242):Coronavirus Proteases
Molecule of the Month (242):Coronavirus Proteases
+Jan 31, 2019. EMDB accession codes are about to change! (news from PDBe EMDB page)
EMDB accession codes are about to change! (news from PDBe EMDB page)
- The allocation of 4 digits for EMDB accession codes will soon come to an end. Whilst these codes will remain in use, new EMDB accession codes will include an additional digit and will expand incrementally as the available range of codes is exhausted. The current 4-digit format prefixed with “EMD-” (i.e. EMD-XXXX) will advance to a 5-digit format (i.e. EMD-XXXXX), and so on. It is currently estimated that the 4-digit codes will be depleted around Spring 2019, at which point the 5-digit format will come into force.
- The EM Navigator/Yorodumi systems omit the EMD- prefix.
Related info.: Q: What is EMD? /
Q: What is EMD? /  ID/Accession-code notation in Yorodumi/EM Navigator
ID/Accession-code notation in Yorodumi/EM Navigator
External links: EMDB Accession Codes are Changing Soon! /
EMDB Accession Codes are Changing Soon! /  Contact to PDBj
Contact to PDBj
+Jul 12, 2017. Major update of PDB
Major update of PDB
- wwPDB released updated PDB data conforming to the new PDBx/mmCIF dictionary.
- This is a major update changing the version number from 4 to 5, and with Remediation, in which all the entries are updated.
- In this update, many items about electron microscopy experimental information are reorganized (e.g. em_software).
- Now, EM Navigator and Yorodumi are based on the updated data.
External links: wwPDB Remediation /
wwPDB Remediation /  Enriched Model Files Conforming to OneDep Data Standards Now Available in the PDB FTP Archive
Enriched Model Files Conforming to OneDep Data Standards Now Available in the PDB FTP Archive
-Yorodumi
Thousand views of thousand structures
- Yorodumi is a browser for structure data from EMDB, PDB, SASBDB, etc.
- This page is also the successor to EM Navigator detail page, and also detail information page/front-end page for Omokage search.
- The word "yorodu" (or yorozu) is an old Japanese word meaning "ten thousand". "mi" (miru) is to see.
Related info.: EMDB /
EMDB /  PDB /
PDB /  SASBDB /
SASBDB /  Comparison of 3 databanks /
Comparison of 3 databanks /  Yorodumi Search /
Yorodumi Search /  Aug 31, 2016. New EM Navigator & Yorodumi /
Aug 31, 2016. New EM Navigator & Yorodumi /  Yorodumi Papers /
Yorodumi Papers /  Jmol/JSmol /
Jmol/JSmol /  Function and homology information /
Function and homology information /  Changes in new EM Navigator and Yorodumi
Changes in new EM Navigator and Yorodumi
 Movie
Movie Controller
Controller















































































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)