[English] 日本語
 Yorodumi
Yorodumi- EMDB-13769: CryoEM structure of Apoferritin at 2.6A from Tundra, 100kV microscope -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-13769 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of Apoferritin at 2.6A from Tundra, 100kV microscope | |||||||||
 Map data Map data | Apoferritin data from Tundra, 100kV microscope with CetaF detector | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Fe / METAL BINDING PROTEIN | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.6 Å | |||||||||
 Authors Authors | Karia D / Hlavenkova H / Koh A / Malinsky M / Yu L / Kotecha A | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2025 Journal: Structure / Year: 2025Title: Sub-3 Å resolution protein structure determination by single-particle cryo-EM at 100 keV. Authors: Dimple Karia / Adrian F Koh / Wen Yang / Victoria I Cushing / Benjamin Basanta / Daniel B Mihaylov / Sagar Khavnekar / Ondřej Vyroubal / Miloš Malínský / Ondřej Sháněl / Vojtěch ...Authors: Dimple Karia / Adrian F Koh / Wen Yang / Victoria I Cushing / Benjamin Basanta / Daniel B Mihaylov / Sagar Khavnekar / Ondřej Vyroubal / Miloš Malínský / Ondřej Sháněl / Vojtěch Doležal / Jürgen Plitzko / Lingbo Yu / Gabriel C Lander / A Radu Aricescu / Basil J Greber / Abhay Kotecha /     Abstract: Cryoelectron microscopy (cryo-EM) has transformed structural biology by providing high-resolution insights into biological macromolecules. We report sub-3 Å resolution structures using the 100 keV ...Cryoelectron microscopy (cryo-EM) has transformed structural biology by providing high-resolution insights into biological macromolecules. We report sub-3 Å resolution structures using the 100 keV Tundra cryo-TEM, equipped with the Falcon C direct electron detector (DED). This system combines advanced optics, extreme-brightness field emission gun (XFEG), and SP-TWIN lens to enhance coherence and resolution. The semi-automated loader reduced contamination and drift, enabling extended data collection, while the high detective quantum efficiency (DQE) of Falcon C improved signal-to-noise ratio. We validated performance by determining structures of biological samples, including apoferritin (2.1 Å), T20S proteasome (2.7 Å), GABA receptor (2.8 Å), hemoglobin (5.0 Å), transthyretin (3.5 Å), and AAV9 capsid (2.8 Å), spanning 50 kDa-3.9 MDa. This work highlights the potential of 100 keV transmission electron microscopes (TEMs) to make cryo-EM more accessible. It sets a precedent for using lower voltage TEMs not only for screening, but also for high-resolution protein structure determination. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13769.map.gz emd_13769.map.gz | 9.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13769-v30.xml emd-13769-v30.xml emd-13769.xml emd-13769.xml | 11.4 KB 11.4 KB | Display Display |  EMDB header EMDB header |
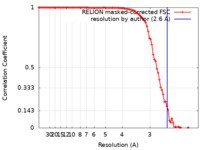
| FSC (resolution estimation) |  emd_13769_fsc.xml emd_13769_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_13769.png emd_13769.png | 215.6 KB | ||
| Filedesc metadata |  emd-13769.cif.gz emd-13769.cif.gz | 4.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13769 http://ftp.pdbj.org/pub/emdb/structures/EMD-13769 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13769 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13769 | HTTPS FTP |
-Validation report
| Summary document |  emd_13769_validation.pdf.gz emd_13769_validation.pdf.gz | 397.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_13769_full_validation.pdf.gz emd_13769_full_validation.pdf.gz | 397.1 KB | Display | |
| Data in XML |  emd_13769_validation.xml.gz emd_13769_validation.xml.gz | 11 KB | Display | |
| Data in CIF |  emd_13769_validation.cif.gz emd_13769_validation.cif.gz | 14.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13769 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13769 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13769 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13769 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10844 (Title: CryoEM structure of Apoferritin at 2.6A from Tundra, 100kV microscope EMPIAR-10844 (Title: CryoEM structure of Apoferritin at 2.6A from Tundra, 100kV microscopeData size: 1.0 TB Data #1: Unaligned multiframe movies of ApoF from CETA-F camera [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_13769.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13769.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Apoferritin data from Tundra, 100kV microscope with CetaF detector | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0476 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Apoferritin
| Entire | Name: Apoferritin |
|---|---|
| Components |
|
-Supramolecule #1: Apoferritin
| Supramolecule | Name: Apoferritin / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 480 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Details: 20mM HEPES pH 7.5 and 300mM NaCl |
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
| Details | 4mg/ml protein in 20mM HEPES pH 7.5 and 300mM NaCl |
- Electron microscopy
Electron microscopy
| Microscope | TFS TUNDRA |
|---|---|
| Image recording | Film or detector model: FEI CETA (4k x 4k) / Number grids imaged: 1 / Average electron dose: 35.0 e/Å2 |
| Electron beam | Acceleration voltage: 100 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 1.6 mm / Nominal magnification: 180000 |
| Sample stage | Specimen holder model: OTHER / Cooling holder cryogen: NITROGEN |
 Movie
Movie Controller
Controller






































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)