[English] 日本語
 Yorodumi
Yorodumi- EMDB-21951: Cryo-EM Structure of Human Apoferritin Light Chain Vitrified Usin... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21951 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM Structure of Human Apoferritin Light Chain Vitrified Using Back-it-up | ||||||||||||
 Map data Map data | Sharpened map | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Human / apoferritin / ferritin / light chain / back-it-up / through-grid wicking / METAL BINDING PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationferritin complex / Scavenging by Class A Receptors / Golgi Associated Vesicle Biogenesis / autolysosome / ferric iron binding / autophagosome / iron ion transport / ferrous iron binding / Iron uptake and transport / azurophil granule lumen ...ferritin complex / Scavenging by Class A Receptors / Golgi Associated Vesicle Biogenesis / autolysosome / ferric iron binding / autophagosome / iron ion transport / ferrous iron binding / Iron uptake and transport / azurophil granule lumen / intracellular iron ion homeostasis / iron ion binding / Neutrophil degranulation / extracellular exosome / extracellular region / identical protein binding / membrane / cytoplasm / cytosol Similarity search - Function | ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.0 Å | ||||||||||||
 Authors Authors | Tan YZ / Rubinstein JL | ||||||||||||
| Funding support |  Canada, 3 items Canada, 3 items
| ||||||||||||
 Citation Citation |  Journal: Acta Crystallogr D Struct Biol / Year: 2020 Journal: Acta Crystallogr D Struct Biol / Year: 2020Title: Through-grid wicking enables high-speed cryoEM specimen preparation. Authors: Yong Zi Tan / John L Rubinstein /  Abstract: Blotting times for conventional cryoEM specimen preparation complicate time-resolved studies and lead to some specimens adopting preferred orientations or denaturing at the air-water interface. Here, ...Blotting times for conventional cryoEM specimen preparation complicate time-resolved studies and lead to some specimens adopting preferred orientations or denaturing at the air-water interface. Here, it is shown that solution sprayed onto one side of a holey cryoEM grid can be wicked through the grid by a glass-fiber filter held against the opposite side, often called the `back', of the grid, producing a film suitable for vitrification. This process can be completed in tens of milliseconds. Ultrasonic specimen application and through-grid wicking were combined in a high-speed specimen-preparation device that was named `Back-it-up' or BIU. The high liquid-absorption capacity of the glass fiber compared with self-wicking grids makes the method relatively insensitive to the amount of sample applied. Consequently, through-grid wicking produces large areas of ice that are suitable for cryoEM for both soluble and detergent-solubilized protein complexes. The speed of the device increases the number of views for a specimen that suffers from preferred orientations. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21951.map.gz emd_21951.map.gz | 85.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21951-v30.xml emd-21951-v30.xml emd-21951.xml emd-21951.xml | 28 KB 28 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_21951_fsc.xml emd_21951_fsc.xml | 11 KB | Display |  FSC data file FSC data file |
| Images |  emd_21951.png emd_21951.png | 198.6 KB | ||
| Masks |  emd_21951_msk_1.map emd_21951_msk_1.map | 91.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-21951.cif.gz emd-21951.cif.gz | 6.3 KB | ||
| Others |  emd_21951_additional_1.map.gz emd_21951_additional_1.map.gz emd_21951_additional_2.map.gz emd_21951_additional_2.map.gz emd_21951_additional_3.map.gz emd_21951_additional_3.map.gz emd_21951_additional_4.map.gz emd_21951_additional_4.map.gz emd_21951_additional_5.map.gz emd_21951_additional_5.map.gz emd_21951_half_map_1.map.gz emd_21951_half_map_1.map.gz emd_21951_half_map_2.map.gz emd_21951_half_map_2.map.gz | 43.9 MB 7.9 MB 194.9 KB 20.5 MB 23.7 MB 84.1 MB 84.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21951 http://ftp.pdbj.org/pub/emdb/structures/EMD-21951 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21951 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21951 | HTTPS FTP |
-Related structure data
| Related structure data |  6wx6MC  6wxbC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10421 (Title: Single-Particle CryoEM of Human Apoferritin Light Chain Vitrified Using Back-it-up EMPIAR-10421 (Title: Single-Particle CryoEM of Human Apoferritin Light Chain Vitrified Using Back-it-upData size: 2.1 TB Data #1: Unaligned Falcon IV movie frames [micrographs - multiframe] Data #2: Aligned Dose-Weighted Micrographs [micrographs - single frame] Data #3: Final Particle Stack with Final Euler Angles and Shifts [picked particles - multiframe - processed]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_21951.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21951.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.816 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_21951_msk_1.map emd_21951_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
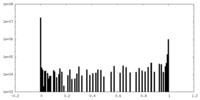
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Raw map
| File | emd_21951_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Raw map | ||||||||||||
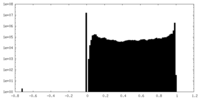
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Local resolution map
| File | emd_21951_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Local resolution map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: 3DFSC - Thresholded and Binarized
| File | emd_21951_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3DFSC - Thresholded and Binarized | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: 3DFSC - Thresholded
| File | emd_21951_additional_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3DFSC - Thresholded | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: 3DFSC - Raw
| File | emd_21951_additional_5.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3DFSC - Raw | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 1
| File | emd_21951_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 2
| File | emd_21951_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Human apoferritin light chain
| Entire | Name: Human apoferritin light chain |
|---|---|
| Components |
|
-Supramolecule #1: Human apoferritin light chain
| Supramolecule | Name: Human apoferritin light chain / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 580 KDa |
-Macromolecule #1: Ferritin light chain
| Macromolecule | Name: Ferritin light chain / type: protein_or_peptide / ID: 1 / Number of copies: 24 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 24.275898 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: TGWSHPQFEK LKGGSSRGGG GGSGGSGGSG GSMSSQIRQN YSTDVEAAVN SLVNLYLQAS YTYLSLGFYF DRDDVALEGV SHFFRELAE EKREGYERLL KMQNQRGGRA LFQDIKKPAE DEWGKTPDAM KAAMALEKKL NQALLDLHAL GSARTDPHLC D FLETHFLD ...String: TGWSHPQFEK LKGGSSRGGG GGSGGSGGSG GSMSSQIRQN YSTDVEAAVN SLVNLYLQAS YTYLSLGFYF DRDDVALEGV SHFFRELAE EKREGYERLL KMQNQRGGRA LFQDIKKPAE DEWGKTPDAM KAAMALEKKL NQALLDLHAL GSARTDPHLC D FLETHFLD EEVKLIKKMG DHLTNLHRLG GPEAGLGEYL FERLTLRHDG GSGGSGGSGG SGGGASGGS UniProtKB: Ferritin light chain |
-Macromolecule #2: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 2 / Number of copies: 8 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Macromolecule #3: water
| Macromolecule | Name: water / type: ligand / ID: 3 / Number of copies: 2420 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Grid | Model: Homemade / Material: COPPER/RHODIUM / Support film - Material: GOLD / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 120 sec. / Pretreatment - Atmosphere: AIR Details: Both sides of the grid were glow discharged for 120 seconds. |
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 50 % / Chamber temperature: 298 K / Instrument: HOMEMADE PLUNGER Details: Back-it-up (ultrasonic specimen application and through-grid wicking in a high-speed specimen preparation device) was used. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 2 / Number real images: 3168 / Average exposure time: 9.0 sec. / Average electron dose: 27.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)