[English] 日本語
 Yorodumi
Yorodumi- EMDB-11603: Human heavy-chain apoferritin refined from tilt series data using M -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11603 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human heavy-chain apoferritin refined from tilt series data using M | ||||||||||||||||||
 Map data Map data | |||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||
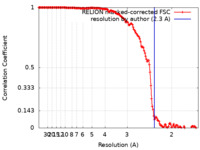
| Method | subtomogram averaging / cryo EM / Resolution: 2.3 Å | ||||||||||||||||||
 Authors Authors | Tegunov D / Xue L / Dienemann C / Cramer P / Mahamid J | ||||||||||||||||||
| Funding support |  Germany, 5 items Germany, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Nat Methods / Year: 2021 Journal: Nat Methods / Year: 2021Title: Multi-particle cryo-EM refinement with M visualizes ribosome-antibiotic complex at 3.5 Å in cells. Authors: Dimitry Tegunov / Liang Xue / Christian Dienemann / Patrick Cramer / Julia Mahamid /  Abstract: Cryo-electron microscopy (cryo-EM) enables macromolecular structure determination in vitro and inside cells. In addition to aligning individual particles, accurate registration of sample motion and ...Cryo-electron microscopy (cryo-EM) enables macromolecular structure determination in vitro and inside cells. In addition to aligning individual particles, accurate registration of sample motion and three-dimensional deformation during exposures are crucial for achieving high-resolution reconstructions. Here we describe M, a software tool that establishes a reference-based, multi-particle refinement framework for cryo-EM data and couples a comprehensive spatial deformation model to in silico correction of electron-optical aberrations. M provides a unified optimization framework for both frame-series and tomographic tilt-series data. We show that tilt-series data can provide the same resolution as frame-series data on a purified protein specimen, indicating that the alignment step no longer limits the resolution obtainable from tomographic data. In combination with Warp and RELION, M resolves to residue level a 70S ribosome bound to an antibiotic inside intact bacterial cells. Our work provides a computational tool that facilitates structural biology in cells. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11603.map.gz emd_11603.map.gz | 139.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11603-v30.xml emd-11603-v30.xml emd-11603.xml emd-11603.xml | 14.1 KB 14.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_11603_fsc.xml emd_11603_fsc.xml | 15.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_11603.png emd_11603.png | 92.7 KB | ||
| Masks |  emd_11603_msk_1.map emd_11603_msk_1.map | 26.2 MB |  Mask map Mask map | |
| Others |  emd_11603_half_map_1.map.gz emd_11603_half_map_1.map.gz emd_11603_half_map_2.map.gz emd_11603_half_map_2.map.gz | 24.2 MB 24.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11603 http://ftp.pdbj.org/pub/emdb/structures/EMD-11603 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11603 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11603 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10491 (Title: Apoferritin tilt series and frame series used to benchmark the resolution of both data types in M EMPIAR-10491 (Title: Apoferritin tilt series and frame series used to benchmark the resolution of both data types in MData size: 39.6 Data #1: Unaligned frame series of apoferritin [micrographs - multiframe] Data #2: Unaligned tilt movies of apoferritin and corresponding MDOC metadata [tilt series]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_11603.map.gz / Format: CCP4 / Size: 149.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11603.map.gz / Format: CCP4 / Size: 149.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.415 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_11603_msk_1.map emd_11603_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_11603_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_11603_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Heavy-chain apoferritin
| Entire | Name: Heavy-chain apoferritin |
|---|---|
| Components |
|
-Supramolecule #1: Heavy-chain apoferritin
| Supramolecule | Name: Heavy-chain apoferritin / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Molecular weight | Experimental: 500 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3.8 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Average electron dose: 2.7 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)