+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20028 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human apoferritin at 3.08 Angstrom | ||||||||||||
 Map data Map data | Human Apoferritin | ||||||||||||
 Sample Sample |
| ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.08 Å | ||||||||||||
 Authors Authors | Zhang K / Pintilie G / Li S / Chiu W | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Methods / Year: 2020 Journal: Nat Methods / Year: 2020Title: Measurement of atom resolvability in cryo-EM maps with Q-scores. Authors: Grigore Pintilie / Kaiming Zhang / Zhaoming Su / Shanshan Li / Michael F Schmid / Wah Chiu /  Abstract: Cryogenic electron microscopy (cryo-EM) maps are now at the point where resolvability of individual atoms can be achieved. However, resolvability is not necessarily uniform throughout the map. We ...Cryogenic electron microscopy (cryo-EM) maps are now at the point where resolvability of individual atoms can be achieved. However, resolvability is not necessarily uniform throughout the map. We introduce a quantitative parameter to characterize the resolvability of individual atoms in cryo-EM maps, the map Q-score. Q-scores can be calculated for atoms in proteins, nucleic acids, water, ligands and other solvent atoms, using models fitted to or derived from cryo-EM maps. Q-scores can also be averaged to represent larger features such as entire residues and nucleotides. Averaged over entire models, Q-scores correlate very well with the estimated resolution of cryo-EM maps for both protein and RNA. Assuming the models they are calculated from are well fitted to the map, Q-scores can be used as a measure of resolvability in cryo-EM maps at various scales, from entire macromolecules down to individual atoms. Q-score analysis of multiple cryo-EM maps of the same proteins derived from different laboratories confirms the reproducibility of structural features from side chains down to water and ion atoms. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20028.map.gz emd_20028.map.gz | 135.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20028-v30.xml emd-20028-v30.xml emd-20028.xml emd-20028.xml | 19.6 KB 19.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_20028.png emd_20028.png | 189.2 KB | ||
| Others |  emd_20028_additional_1.map.gz emd_20028_additional_1.map.gz emd_20028_additional_2.map.gz emd_20028_additional_2.map.gz emd_20028_half_map_1.map.gz emd_20028_half_map_1.map.gz emd_20028_half_map_2.map.gz emd_20028_half_map_2.map.gz | 108.9 MB 837.8 KB 109.3 MB 109.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20028 http://ftp.pdbj.org/pub/emdb/structures/EMD-20028 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20028 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20028 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_20028.map.gz / Format: CCP4 / Size: 144.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20028.map.gz / Format: CCP4 / Size: 144.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Human Apoferritin | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.65 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
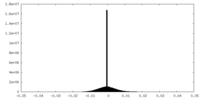
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: unmasked map
| File | emd_20028_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unmasked map | ||||||||||||
| Projections & Slices |
| ||||||||||||
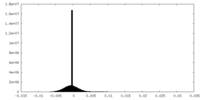
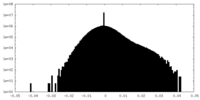
| Density Histograms |
-Additional map: One protomer
| File | emd_20028_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | One protomer | ||||||||||||
| Projections & Slices |
| ||||||||||||
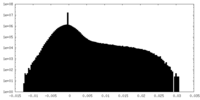
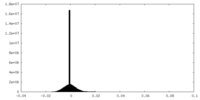
| Density Histograms |
-Half map: Half map 1
| File | emd_20028_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
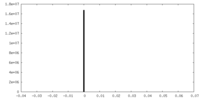
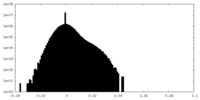
| Density Histograms |
-Half map: Half map 2
| File | emd_20028_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
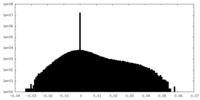
| Density Histograms |
- Sample components
Sample components
-Entire : Human Apoferritin
| Entire | Name: Human Apoferritin |
|---|---|
| Components |
|
-Supramolecule #1: Human Apoferritin
| Supramolecule | Name: Human Apoferritin / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Molecular weight | Experimental: 480 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.5 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Details: unspecified |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 1-30 / Number grids imaged: 1 / Number real images: 1100 / Average exposure time: 6.0 sec. / Average electron dose: 7.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated defocus max: 1.5 µm / Calibrated defocus min: 0.4 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal magnification: 215000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)