[English] 日本語
 Yorodumi
Yorodumi- EMDB-11774: Cryo-EM structure of the signal sequence-engaged post-translation... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11774 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the signal sequence-engaged post-translational Sec translocon | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Post-translational translocation / Protein translocation / Sec complex / Signal sequence / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationmating pheromone activity / mating / misfolded protein transport / Sec62/Sec63 complex / translocon complex / Insertion of tail-anchored proteins into the endoplasmic reticulum membrane / pheromone-dependent signal transduction involved in conjugation with cellular fusion / rough endoplasmic reticulum membrane / cytosol to endoplasmic reticulum transport / Ssh1 translocon complex ...mating pheromone activity / mating / misfolded protein transport / Sec62/Sec63 complex / translocon complex / Insertion of tail-anchored proteins into the endoplasmic reticulum membrane / pheromone-dependent signal transduction involved in conjugation with cellular fusion / rough endoplasmic reticulum membrane / cytosol to endoplasmic reticulum transport / Ssh1 translocon complex / Sec61 translocon complex / protein-transporting ATPase activity / post-translational protein targeting to endoplasmic reticulum membrane / SRP-dependent cotranslational protein targeting to membrane, translocation / filamentous growth / signal sequence binding / SRP-dependent cotranslational protein targeting to membrane / post-translational protein targeting to membrane, translocation / cupric ion binding / peptide transmembrane transporter activity / nuclear inner membrane / retrograde protein transport, ER to cytosol / protein transmembrane transporter activity / ERAD pathway / guanyl-nucleotide exchange factor activity / cell periphery / ribosome binding / endoplasmic reticulum membrane / structural molecule activity / endoplasmic reticulum / mitochondrion / extracellular region / membrane / cytosol Similarity search - Function | |||||||||
| Biological species |  | |||||||||
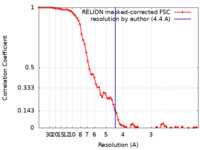
| Method | single particle reconstruction / cryo EM / Resolution: 4.4 Å | |||||||||
 Authors Authors | Weng T-H / Beatrix B | |||||||||
 Citation Citation |  Journal: EMBO J / Year: 2021 Journal: EMBO J / Year: 2021Title: Architecture of the active post-translational Sec translocon. Authors: Tsai-Hsuan Weng / Wieland Steinchen / Birgitta Beatrix / Otto Berninghausen / Thomas Becker / Gert Bange / Jingdong Cheng / Roland Beckmann /  Abstract: In eukaryotes, most secretory and membrane proteins are targeted by an N-terminal signal sequence to the endoplasmic reticulum, where the trimeric Sec61 complex serves as protein-conducting channel ...In eukaryotes, most secretory and membrane proteins are targeted by an N-terminal signal sequence to the endoplasmic reticulum, where the trimeric Sec61 complex serves as protein-conducting channel (PCC). In the post-translational mode, fully synthesized proteins are recognized by a specialized channel additionally containing the Sec62, Sec63, Sec71, and Sec72 subunits. Recent structures of this Sec complex in the idle state revealed the overall architecture in a pre-opened state. Here, we present a cryo-EM structure of the yeast Sec complex bound to a substrate, and a crystal structure of the Sec62 cytosolic domain. The signal sequence is inserted into the lateral gate of Sec61α similar to previous structures, yet, with the gate adopting an even more open conformation. The signal sequence is flanked by two Sec62 transmembrane helices, the cytoplasmic N-terminal domain of Sec62 is more rigidly positioned, and the plug domain is relocated. We crystallized the Sec62 domain and mapped its interaction with the C-terminus of Sec63. Together, we obtained a near-complete and integrated model of the active Sec complex. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11774.map.gz emd_11774.map.gz | 28.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11774-v30.xml emd-11774-v30.xml emd-11774.xml emd-11774.xml | 31.2 KB 31.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_11774_fsc.xml emd_11774_fsc.xml | 7.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_11774.png emd_11774.png | 51.9 KB | ||
| Filedesc metadata |  emd-11774.cif.gz emd-11774.cif.gz | 8.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11774 http://ftp.pdbj.org/pub/emdb/structures/EMD-11774 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11774 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11774 | HTTPS FTP |
-Validation report
| Summary document |  emd_11774_validation.pdf.gz emd_11774_validation.pdf.gz | 570.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_11774_full_validation.pdf.gz emd_11774_full_validation.pdf.gz | 569.8 KB | Display | |
| Data in XML |  emd_11774_validation.xml.gz emd_11774_validation.xml.gz | 9.6 KB | Display | |
| Data in CIF |  emd_11774_validation.cif.gz emd_11774_validation.cif.gz | 12.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11774 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11774 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11774 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11774 | HTTPS FTP |
-Related structure data
| Related structure data |  7aftMC  6zzzC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_11774.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11774.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.059 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
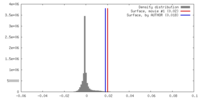
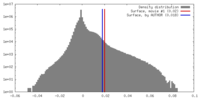
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : Signal sequence-engaged post-translational Sec translocon
+Supramolecule #1: Signal sequence-engaged post-translational Sec translocon
+Supramolecule #2: Signal sequence-engaged post-translational Sec translocon
+Supramolecule #3: Translocation protein SEC62
+Supramolecule #4: Mating factor alpha-1
+Macromolecule #1: Protein transport protein SEC61
+Macromolecule #2: Protein transport protein SBH1
+Macromolecule #3: Protein transport protein SSS1
+Macromolecule #4: Protein translocation protein SEC63
+Macromolecule #5: Translocation protein SEC66
+Macromolecule #6: Translocation protein SEC72
+Macromolecule #7: Translocation protein SEC62,Translocation protein SEC62
+Macromolecule #8: Mating factor alpha-1,Mating factor alpha-1
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| ||||||||||||||||||
| Grid | Model: UltrAuFoil R2/2 / Material: GOLD | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | #0 - Image recording ID: 1 / #0 - Film or detector model: GATAN K2 QUANTUM (4k x 4k) / #0 - Detector mode: COUNTING / #0 - Number grids imaged: 1 / #0 - Average electron dose: 36.0 e/Å2 / #1 - Image recording ID: 2 / #1 - Film or detector model: GATAN K2 QUANTUM (4k x 4k) / #1 - Detector mode: COUNTING / #1 - Number grids imaged: 1 / #1 - Average electron dose: 48.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)