[English] 日本語
 Yorodumi
Yorodumi- EMDB-11581: Outer Dynein Arm-Shulin complex - Dyh3 motor region (Tetrahymena ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11581 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Outer Dynein Arm-Shulin complex - Dyh3 motor region (Tetrahymena thermophila) | |||||||||
 Map data Map data | Main map of Dyh3 motor region (resampled on a full-length ODA-Shulin structure from a smaller subset of particles using EMDA). -135 angstrom auto-sharpened map. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Cilia / dynein / microtubules / motor / MOTOR PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationaxonemal dynein complex / minus-end-directed microtubule motor activity / dynein light intermediate chain binding / microtubule-based movement / dynein intermediate chain binding / microtubule / ATP binding Similarity search - Function | |||||||||
| Biological species |   Tetrahymena thermophila (strain SB210) (eukaryote) Tetrahymena thermophila (strain SB210) (eukaryote) | |||||||||
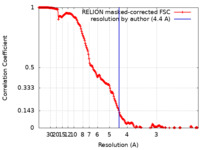
| Method | single particle reconstruction / cryo EM / Resolution: 4.4 Å | |||||||||
 Authors Authors | Mali GR / Abid Ali F / Lau CK / Carter AP | |||||||||
| Funding support |  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2021 Journal: Science / Year: 2021Title: Shulin packages axonemal outer dynein arms for ciliary targeting. Authors: Girish R Mali / Ferdos Abid Ali / Clinton K Lau / Farida Begum / Jérôme Boulanger / Jonathan D Howe / Zhuo A Chen / Juri Rappsilber / Mark Skehel / Andrew P Carter /   Abstract: The main force generators in eukaryotic cilia and flagella are axonemal outer dynein arms (ODAs). During ciliogenesis, these ~1.8-megadalton complexes are assembled in the cytoplasm and targeted to ...The main force generators in eukaryotic cilia and flagella are axonemal outer dynein arms (ODAs). During ciliogenesis, these ~1.8-megadalton complexes are assembled in the cytoplasm and targeted to cilia by an unknown mechanism. Here, we used the ciliate to identify two factors (Q22YU3 and Q22MS1) that bind ODAs in the cytoplasm and are required for ODA delivery to cilia. Q22YU3, which we named Shulin, locked the ODA motor domains into a closed conformation and inhibited motor activity. Cryo-electron microscopy revealed how Shulin stabilized this compact form of ODAs by binding to the dynein tails. Our findings provide a molecular explanation for how newly assembled dyneins are packaged for delivery to the cilia. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11581.map.gz emd_11581.map.gz | 216.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11581-v30.xml emd-11581-v30.xml emd-11581.xml emd-11581.xml | 27.5 KB 27.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_11581_fsc.xml emd_11581_fsc.xml | 18.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_11581.png emd_11581.png | 65.6 KB | ||
| Masks |  emd_11581_msk_1.map emd_11581_msk_1.map emd_11581_msk_2.map emd_11581_msk_2.map | 512 MB 347.6 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-11581.cif.gz emd-11581.cif.gz | 9.9 KB | ||
| Others |  emd_11581_additional_1.map.gz emd_11581_additional_1.map.gz emd_11581_additional_2.map.gz emd_11581_additional_2.map.gz emd_11581_half_map_1.map.gz emd_11581_half_map_1.map.gz emd_11581_half_map_2.map.gz emd_11581_half_map_2.map.gz | 216.4 MB 216.4 MB 414.4 MB 414.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11581 http://ftp.pdbj.org/pub/emdb/structures/EMD-11581 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11581 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11581 | HTTPS FTP |
-Related structure data
| Related structure data |  6zyyMC  6zywC  6zyxC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_11581.map.gz / Format: CCP4 / Size: 347.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11581.map.gz / Format: CCP4 / Size: 347.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Main map of Dyh3 motor region (resampled on a full-length ODA-Shulin structure from a smaller subset of particles using EMDA). -135 angstrom auto-sharpened map. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.11 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
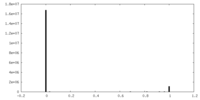
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_11581_msk_1.map emd_11581_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
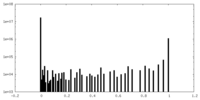
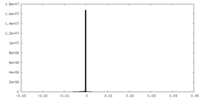
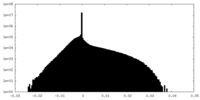
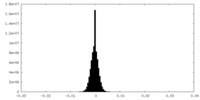
| Density Histograms |
-Mask #2
| File |  emd_11581_msk_2.map emd_11581_msk_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
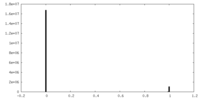
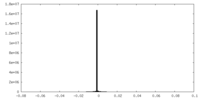
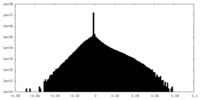
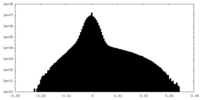
| Density Histograms |
-Additional map: Additional map of Dyh3 motor region (resampled on...
| File | emd_11581_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Additional map of Dyh3 motor region (resampled on a full-length ODA-Shulin structure from a smaller subset of particles using EMDA). -75 angstrom manually sharpened map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Additional map of Dyh3 motor region (resampled on...
| File | emd_11581_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Additional map of Dyh3 motor region (resampled on a full-length ODA-Shulin structure from a smaller subset of particles using EMDA). -200 angstrom manually sharpened map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
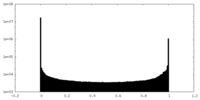
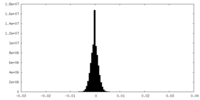
| Density Histograms |
-Half map: Half map 2 of Dyh3 motor region map (Z-flipped, non-resampled)
| File | emd_11581_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 of Dyh3 motor region map (Z-flipped, non-resampled) | ||||||||||||
| Projections & Slices |
| ||||||||||||
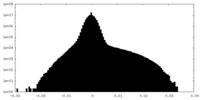
| Density Histograms |
-Half map: Half map 1 of Dyh3 motor region map (Z-flipped, non-resampled)
| File | emd_11581_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 of Dyh3 motor region map (Z-flipped, non-resampled) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Tetrahymena thermophila ODA Dyh3 motor region (includes Shulin C3...
| Entire | Name: Tetrahymena thermophila ODA Dyh3 motor region (includes Shulin C3 finger) |
|---|---|
| Components |
|
-Supramolecule #1: Tetrahymena thermophila ODA Dyh3 motor region (includes Shulin C3...
| Supramolecule | Name: Tetrahymena thermophila ODA Dyh3 motor region (includes Shulin C3 finger) type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 1.8 MDa |
-Macromolecule #1: Dynein heavy chain, outer arm protein
| Macromolecule | Name: Dynein heavy chain, outer arm protein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Tetrahymena thermophila (strain SB210) (eukaryote) Tetrahymena thermophila (strain SB210) (eukaryote)Strain: SB210 |
| Molecular weight | Theoretical: 534.328812 KDa |
| Sequence | String: MLSSKYSKRI AWMKTTICDS LQLKDMIVEE SFQYEKNKNL LEQFLSGEGL NKIFAYYQVQ EQAQNDDIKD TGAQDPVLFF TTGDLEKIQ DKAVWFLRIT NPADDKKKAS QQDGNDNDII FGEITPNTVP MLNALMESVY SRQIDHIITE KIQFWGVAEE E QVLEFQQH ...String: MLSSKYSKRI AWMKTTICDS LQLKDMIVEE SFQYEKNKNL LEQFLSGEGL NKIFAYYQVQ EQAQNDDIKD TGAQDPVLFF TTGDLEKIQ DKAVWFLRIT NPADDKKKAS QQDGNDNDII FGEITPNTVP MLNALMESVY SRQIDHIITE KIQFWGVAEE E QVLEFQQH SNKFSSEVRE AINLMSPGTE HFKLDYEAIS GLSESEKMQH YEMKFNEWIN LISSQLNDDS EVRKDEKDAG PA TELIYWR SRMQKITNWS EQLKSKDFQI VKASLQRHKN HDNQRPRGDE SLSKLMMEYN RLDLLLTDKL NEAKDNVKYL TTL EKFIEP LYNGTPQQII DTLPALMNAI KMIHTIARFY NTTDKMTGLF IKITNQMIKN CKDRILNKKD NGDNPSLYKM IWEQ DPAEL IEVLGSCIKL YCEYKKCYND TKEKVADMPK GKTFDFSDAQ IFGKFDTFVR RLQKLIEIFS NIQQFNALAK HNLEG MDVL TNKFKKIIDD FKKKGHNLLD TANNKFDRDW VEFNVEISHL DGELQNFIDN NFNRFRNIEY SLKLLHKFQS TIKRDS LKH NLTSRYNAIL HNYATELDTI QRVFQDQKSN PPLVRNMPPE AGKIIWARHL FQKITGPINI FPENVINSTE IRRYYGS YN TLGKQLTIYE MWFYQDWVNK IEQSKAALQA TLIVRHDENK KLYVNFDLEI MQLIREAKCL DRQGIEIPES ARIILLQE D KFKTYYNELL YALKEYERIN SKIKPICKNL LLPHIEDLDL KLRPGMVTLT WTSMNIESYL YYVHQGLKKL EQLIINVND IIENRIENNL KTVSKVVLVH LPQDTKPLSL DSFVQLQEEY INSKTDFLTS KNVEVERAVD DLLQTIMLYP LDPHVDPVLP EETKRIKRY YFWYFYQALL NSTQNSLNAM KYRVCGKKIP GANTLQNLKP FFQVEVQLNG DKVTLNPSLQ EIQKSINRAA T AVLRCSKH LYNWDQQNKD STDKATFYDM IACDKEIVKV ILLLTGSIQG TKNKVNEFLS GFTKFEWLCK ESIQESIKKF SK NGPTLQN YEDQLKKFSQ IEEEIEKIVP TYKIGAMELM THNICTSLST WAKEWKLQYS QDLHKRARQL LDSLTEQTKM LST KLSKPV KDIDSLGYVM ETLEQIRKEQ AEIDMKFNPV QEMYSLLDNY LPGGITDKDE MDARSLLRRN WDILIQQAEI KGKE YQHKQ AIYLKELKQS IKDFTNQVSI FRRDYEKNGP MVEGISPAEA MERLRRFEDE YDVKYQMYKI NARGENLFGL QNQKY PELE KTDAEIKNLN KLYNLYDSVI KNIQQFKEKS WQDVSKDDLA KMEEDAGKYG EQCSRLPKDL KEWQAYRDLK NYIDSL REQ LPLIISLKKP SIMPRHWEKI KEITNTKLNY ENPDQFYIEE IMGAKLLDFR EDIEDITESA DKQLKIRTGL DEINLYW ND MQFQFGIWGK RDVPCMLNGL IVGTILERLE EDQLQLSTFN SQRHVTPFKA EVENLIRTFS DVNDTLDMWV KVQKLWTS L EPVFTGGDIA RQMPLQAKQF QGIDKNWMKI MEKAVETKKV IPCCQNDMLK DFLPDLNRKL EDCQKMLEAY LEGKRKKFP RFYFVSNPTL LKILSQGSEP TSIQEDFEKL FDAITKVTFE SAKDKKNPAL KQITQIQQVI GRNEENISLT GYYVKCEGNI EDWLKKLEQ NMQQTLKDIA SAAAQQVFQV GLKEFVSSQA SQIALLGLQI LWTSKVNEGL ERLSRNERNA MDIKRNEIKE H MNILSSMC LEDLNGAVER TKVETLVTIQ VHQKDISMDL KCKDVNDFEW QKQTRIAWKT DIDECIISIT DWDSPYSYEF LG AKERLCI TPLTDRCYIT LAQAMSMYYG GAPAGPAGTG KTETVKDLGR TLGVFVVVTN CSDQHRYRDM AKIFKGLVQS GLW GCFDEF NRIDLEVLSV VAMQVESITT ARKQHMKKFM FPEEEIEIEL IPTVSYFITM NPGYAGRQEL PENLKVLFRG VSMM VPDRE IIIKVKLASV GYLQIDLLAK KFNVLYRLCE EQLSKQRHYD FGLRNILSVL RTAGNTKRQE IKSDEEMLLM RSLRD MNLS KLVADDIPLF NGLLADIFPK LKEVPKKLYP DVEKKIPEEI NAESYLINTP SFQLKIIQLY ETCLVRHGFM LVGPTG SGK STIMKILTEV LTKLGSPHKI VIMNPKAITA EQMYGVKSEI SDDWIPGVFS TIWAKSNNRA LKHTTWITCD GPVDAIW IE NLNTVLDDNK ILTLANGERI AMTENCKVVF EVENLNNASP ATVSRCGQVY VSPTDLGYEA VIEGWIRNRK ASGRAEES D KLGNILRKYL INMRFIELQS KECKEPMMDT SPVISVINIL NLLTGCLQYF VQTQRTLSEQ EYEKFIVYSM AWAIGGIYE AQDRVRFHEL LLAKNAPIPQ KGKENETVFD YYVSQDYLDW KICSPEEWVP PQSLQFSQLL LPTLDSFRAE MLLNFILTQP KSHTCSNSA LLIGGSGTAK TSSVLLYCNK FDPQKMLFKR TNFSSATSPF MFQSTIEAEC DFKVGKEFAP PGNKMMTIFI D DMSMPFVN KWGDQITLEL VRQLIETGGF YMLDKTQRGN QRKMKNLQYI GAMNHPGGGR NDIPNRLKRQ FFIFNMILPL SI EGIYGPI IKHMFKQKYF SDSTYKVIES LTSATIALWN KVKSTMLPTP AKFHYVFNMR ELSRIFKGIL TCKKDTINDA PKS MKIKPE LFLVGLWRHE AERVLADKLV NNKDKDTVMG YIQEVSLESF SQIENEILEK YSSEKTFLFC DFLRPDVINE DGII EEEAP KIYEAIDSLT ELRKRCNFLL SFYNDRNPSK KMPLVLFDDA LKHLLRISRI IRQPRSSGLL VGVGGSGKQS LTRLA GFIG KNLIQQIIVT KTYSDKDLKE DIKKGFDDAG HLGKQVTFLM TDSEVKKEEF LEYINMVLST GEIPNLLAKD EREVWL GDI SQAYCKEKNL GNIDPPQSEL WTYFVDRVRD NFHIMLCFSP VGQKFRERAR KFPALFNECT IDWFLPWPEE ALVSVAE TF IKNFDKLDTK EETKQELMKH MGNVHLMVNE ICDEYYQKMR RQVYVTPKSF LSYLNSYKTL YIEKYDELDQ QEESFKIG L NKIQEATITI NQMEISLKEE EIQLNEATEK TNQLLANLDK ESKKANQKGE EVAATNKQCE IQAEQISKEK EEAERELEA ALPALRRAQE AVDSIESKDI VELKANKKPL DIIKYIMDAV LVFFKARLIP IQIEERVFNK KEGKAVLFLK ESYDESGIQT LGDMNFMKK LKEFEKDSIN EETIELLEPY LNQSEDWFND TFATKASKAA AGILKWAFAI YEYHQKSKIV KPKRIQVAIA E GRQAIALK ELEKAREDLA QIQAYIKNLK DVYTKQMEEK NELEMKAAKT KKKINTARTL ITSLSGEKDR WGKGAQDISD QK RKLVGNV SLSTAFISYC GPFNAEYRNK LAQQRFVVDM KKRGVPVTPG LELTSFLIDD ATIGEWNLQG LPKDDLSIQN GIM VTNSAR YPLFIDPQGQ GQNWIRNKLS ASIIPERCIT TLSHPKFKDM FLKYCMESGL TLIVENIENE VDPMMDPVLE RQII VKGKT QFVNVAGTEM ELSKEFKLFM TCRLANPSFS PELSAKTTII DFTVTQSGLE QQLLGKVISK EQKALEDSLN QLLAD VNQN QKDLQRLDKN LLERLINSQG NLLDDTELMD VLNNTKTQAK EVAAKLIDAE IKTKEINEKR EQYRPVAIRG SAIYFT MIE VSLVNWMYNS SLEQFLKLFI ESIDLSEKAQ LPSNRVKNII SFLTFHVYRY VNRGLFEKDK ITFILMMAFK ILTTAGT IS SGDVSLFLKS GDALDIKSER QKQISYLEDN QWLNILALSK HTFSGQTLPF FKELPDLISR SENQWRNWID KNDPENFP I PDFAESINQE KEIGSFISLC LVRSLRNDRT LIATQNFISN VLGKEFTDPI SYPIEGIWQE SSNMDPVLFL LSAGADPTS SIDELAKKKK KFPCEKVSMG EGQERVARQV IMKGFVEGGW VILQNCHLGL KFMEEIETLV SPINQIHEDF RLWITCEQHP KFPLGLLQK TLKVTNEPPK GLKAGLYKTF TTIITQEFID KVDHSNWRSL IFTICFLHSI VIERKKFGPL GWCVPYEYNY S DLEASLLY IEKYLTNLMS TPQPNSHNLP ISMNVVRYMI CEVQYGGRIT DDLDRELFIT YGETYLKDGI FGNDYFFYDI MV DGSGQKF KYRIPQNPSA ELIKYQEYIA KVPTVDNPEV FGLHSNADLT FRLKESKEMI NTVMETRPKD SSVGGGKTRE EIV QDKAKD MLKNLPPDYN DVEVRELVSK LGGPNPKTST ERGMTVPLNI FLYQEVTRMQ RVIGLVRKTL QDTILAIDGQ IIMT PEILE AINAIYDAKV PNSWLYDPSG AEISWLLPNL GSWSTSLSDR NKQLNDWLRS GQRPILFWLT GFFNPQGFLT GMKQE VTRN HKKGDGKGGE AWSLDDVVYS TTVKEREKEK DIEQPPAEGV YIKGLYLEGC KWSKNGLDDS DPKKIFADLP ILHVSA INK KKTNEQDRMS NTYLCPVYKY PKRTDKYLIF RVGLPCEGSN NPSHWKLRGV ALLCSTE UniProtKB: Dynein heavy chain, outer arm protein |
-Macromolecule #2: Shulin
| Macromolecule | Name: Shulin / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Tetrahymena thermophila (strain SB210) (eukaryote) Tetrahymena thermophila (strain SB210) (eukaryote)Strain: SB210 |
| Molecular weight | Theoretical: 139.935781 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MFNFFSSANI NQNIPKYSVN DFVFRLKKIE KIVVKEGLDG FLLINGVDSR ENTEYVKLTN WLFLGNSGLE IEENEYLNQI YSDMIVLIK KGTTHIFIDP EALNSLQTLI YSIPNVDVFC PTEKQYEDKD EMELLKMAFF LRVMKPTKKV GILLGQKDKG K INSIEKWP ...String: MFNFFSSANI NQNIPKYSVN DFVFRLKKIE KIVVKEGLDG FLLINGVDSR ENTEYVKLTN WLFLGNSGLE IEENEYLNQI YSDMIVLIK KGTTHIFIDP EALNSLQTLI YSIPNVDVFC PTEKQYEDKD EMELLKMAFF LRVMKPTKKV GILLGQKDKG K INSIEKWP LIQSYGLEEL GVGFFSMNHE VVDLTLRLNA VYKNYDKFFV SKLIYVVAKR LTGHFNSAAG QLGDMKMHKR NL ATESQLT EIFRDTYEIE EISKWVQIRG VNAALPKPRV LFGKNTSADC SKEPSVAPLK DLKYSETFHS FHATFETFDL RTC LRAART YFLAKGVKEE RNLITLNDDE GVPQGYELNI DENQQYKDQD FLANLYLSII IGFNEVMQLI TKDYKNMTEE FIQD YIFQK VSKVYAGFQI PESEITLDKI QIILKAYNSF GEEVKIDFKD TISFKLTPYF FMVRIEQKNI KSQILNNTVL GSLVF AESF ILQEGCYLLL TKEIPYFDLW NCQNDYSEKI EKMKKRILWE PLGKQISDEL PKNRIFVQTG RKSNYGFDIP IMQASY YMH ELGLRIETQR LGWFILFFKE MKEIQITQKM NHTWLIFKVD SNITFNSISK DTIALEFTGD ALEQSFFKIK NYFEENQ IK YEYQVDIPAI FQESQIAKKQ ILNQQSQGQK LITMNSIQNE QFFISYIESK QLMILNQMKD LKLSAYKNLY EQMQISQA I TPVENHIGVI LVNGSYCSGK RKFAENLIRF GSDNNLRLHL YKFDLNEMSE LTEKSYLSGL LKFASEKKIQ NTDVIVASV PHFINTKILI DYFSKSEKIS NAFYIRTIAT KININNIYSN FNKNPVNNVF TYGVEGYSQF LLLDTYNNYD ADVNALNKTL SGVLPGAKI YKIMNNILNP ALAKDILTSI TFISEQNNLN RLKYSVQYDL LTSNGPSSVV FIPFKLPILR EKIRDLIYKK I LQNGNQTL VDTIEAEQKI AEFKELNKNS KDPLMIEIIK LKEKIEIQNA QTSDQAIKID YVKGILRYDS KLKEGLEEIT IT PNYFIER TVKGVDAKEF TEELNGVSFK NVKYTGITNS IINDMGFVFA GKNLNKEKLL ELLYKLVKPL NKQKLRQRKD LTE EEIVDI QFRNRGEGLE NGEFYDGQFW RNIQGLILPH HPKKDEFIEE YLKQEEVRIN QINEQLQQEW ETWKQVYDKI HLDK UniProtKB: Dynein axonemal assembly factor 9 N-terminal domain-containing protein |
-Macromolecule #3: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 3 / Number of copies: 3 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Macromolecule #4: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 4 / Number of copies: 1 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.2 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Grid | Model: Quantifoil R2/2 / Material: GOLD / Support film - Material: GRAPHENE OXIDE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 52.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-6zyy: |
 Movie
Movie Controller
Controller




















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)