[English] 日本語
 Yorodumi
Yorodumi- EMDB-10837: Rix1-Rea1 pre-60S particle - Rea1, body 3 (rigid body refinement,... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10837 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Rix1-Rea1 pre-60S particle - Rea1, body 3 (rigid body refinement, composite structure of Rea1 ring and tail) | |||||||||
 Map data Map data | Local resolution filtered composite map of the Rea1 Ring and Tail of the nucleoplasmic Rix1-Rea1 pre-60S particle | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | pre-60S / Biogenesis / LSU / Large subunit / ribosome assembly / RIBOSOME | |||||||||
| Function / homology |  Function and homology information Function and homology informationprotein-RNA complex remodeling / regulation of ribosomal subunit export from nucleus / preribosome, large subunit precursor / ribosomal large subunit export from nucleus / rRNA processing / ribosomal large subunit assembly / nucleolus / ATP hydrolysis activity / mitochondrion / nucleoplasm ...protein-RNA complex remodeling / regulation of ribosomal subunit export from nucleus / preribosome, large subunit precursor / ribosomal large subunit export from nucleus / rRNA processing / ribosomal large subunit assembly / nucleolus / ATP hydrolysis activity / mitochondrion / nucleoplasm / ATP binding / nucleus Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.2 Å | |||||||||
 Authors Authors | Kater L / Beckmann R | |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2020 Journal: Mol Cell / Year: 2020Title: Construction of the Central Protuberance and L1 Stalk during 60S Subunit Biogenesis. Authors: Lukas Kater / Valentin Mitterer / Matthias Thoms / Jingdong Cheng / Otto Berninghausen / Roland Beckmann / Ed Hurt /  Abstract: Ribosome assembly is driven by numerous assembly factors, including the Rix1 complex and the AAA ATPase Rea1. These two assembly factors catalyze 60S maturation at two distinct states, triggering ...Ribosome assembly is driven by numerous assembly factors, including the Rix1 complex and the AAA ATPase Rea1. These two assembly factors catalyze 60S maturation at two distinct states, triggering poorly understood large-scale structural transitions that we analyzed by cryo-electron microscopy. Two nuclear pre-60S intermediates were discovered that represent previously unknown states after Rea1-mediated removal of the Ytm1-Erb1 complex and reveal how the L1 stalk develops from a pre-mature nucleolar to a mature-like nucleoplasmic state. A later pre-60S intermediate shows how the central protuberance arises, assisted by the nearby Rix1-Rea1 machinery, which was solved in its pre-ribosomal context to molecular resolution. This revealed a Rix1-Ipi3 tetramer anchored to the pre-60S via Ipi1, strategically positioned to monitor this decisive remodeling. These results are consistent with a general underlying principle that temporarily stabilized immature RNA domains are successively remodeled by assembly factors, thereby ensuring failsafe assembly progression. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10837.map.gz emd_10837.map.gz | 4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10837-v30.xml emd-10837-v30.xml emd-10837.xml emd-10837.xml | 23.8 KB 23.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_10837.png emd_10837.png | 66.2 KB | ||
| Filedesc metadata |  emd-10837.cif.gz emd-10837.cif.gz | 9.1 KB | ||
| Others |  emd_10837_additional_1.map.gz emd_10837_additional_1.map.gz emd_10837_additional_2.map.gz emd_10837_additional_2.map.gz | 37.9 MB 36.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10837 http://ftp.pdbj.org/pub/emdb/structures/EMD-10837 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10837 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10837 | HTTPS FTP |
-Related structure data
| Related structure data |  6ylfMC  6yleC  6ylgC  6ylhC  6ylxC  6ylyC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10837.map.gz / Format: CCP4 / Size: 163.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10837.map.gz / Format: CCP4 / Size: 163.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Local resolution filtered composite map of the Rea1 Ring and Tail of the nucleoplasmic Rix1-Rea1 pre-60S particle | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.059 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Local resolution filtered map of the Tail of...
| File | emd_10837_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Local resolution filtered map of the Tail of Rea1 of the nucleoplasmic Rix1-Rea1 pre-60S particle | ||||||||||||
| Projections & Slices |
| ||||||||||||
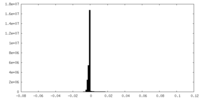
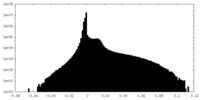
| Density Histograms |
-Additional map: Local resolution filtered map of the Ring of...
| File | emd_10837_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Local resolution filtered map of the Ring of Rea1 of the nucleoplasmic Rix1-Rea1 pre-60S particle | ||||||||||||
| Projections & Slices |
| ||||||||||||
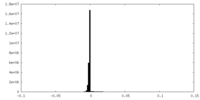
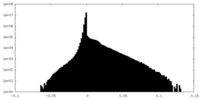
| Density Histograms |
- Sample components
Sample components
-Entire : Rix1-Rea1 pre-60S assembly particle - Rea1 with the Rsa4-UBL domain
| Entire | Name: Rix1-Rea1 pre-60S assembly particle - Rea1 with the Rsa4-UBL domain |
|---|---|
| Components |
|
-Supramolecule #1: Rix1-Rea1 pre-60S assembly particle - Rea1 with the Rsa4-UBL domain
| Supramolecule | Name: Rix1-Rea1 pre-60S assembly particle - Rea1 with the Rsa4-UBL domain type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Rix1-TAP Flag-Rea1 derived pre-60S assembly complex |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Midasin
| Macromolecule | Name: Midasin / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 559.951438 KDa |
| Sequence | String: MSQDRILLDL DVVNQRLILF NSAFPSDAIE APFHFSNKES TSENLDNLAG TILHSRSITG HVFLYKHIFL EIVARWIKDS KKKDYVLVI EKLASIITIF PVAMPLIEDY LDKENDHFIT ILQNPSTQKD SDMFKILLAY YRLLYHNKEV FARFIQPDIL Y QLVDLLTK ...String: MSQDRILLDL DVVNQRLILF NSAFPSDAIE APFHFSNKES TSENLDNLAG TILHSRSITG HVFLYKHIFL EIVARWIKDS KKKDYVLVI EKLASIITIF PVAMPLIEDY LDKENDHFIT ILQNPSTQKD SDMFKILLAY YRLLYHNKEV FARFIQPDIL Y QLVDLLTK EQENQVVIFL ALKVLSLYLD MGEKTLNDML DTYIKSRDSL LGHFEGDSGI DYSFLELNEA KRCANFSKLP SV PECFTIE KKSSYFIIEP QDLSTKVASI CGVIVPKVHT IHDKVFYPLT FVPTHKTVSS LRQLGRKIQN STPIMLIGKA GSG KTFLIN ELSKYMGCHD SIVKIHLGEQ TDAKLLIGTY TSGDKPGTFE WRAGVLATAV KEGRWVLIED IDKAPTDVLS ILLS LLEKR ELTIPSRGET VKAANGFQLI STVRINEDHQ KDSSNKIYNL NMIGMRIWNV IELEEPSEED LTHILAQKFP ILTNL IPKL IDSYKNVKSI YMNTKFISLN KGAHTRVVSV RDLIKLCERL DILFKNNGIN KPDQLIQSSV YDSIFSEAAD CFAGAI GEF KALEPIIQAI GESLDIASSR ISLFLTQHVP TLENLDDSIK IGRAVLLKEK LNIQKKSMNS TLFAFTNHSL RLMEQIS VC IQMTEPVLLV GETGTGKTTV VQQLAKMLAK KLTVINVSQQ TETGDLLGGY KPVNSKTVAV PIQENFETLF NATFSLKK N EKFHKMLHRC FNKNQWKNVV KLWNEAYKMA QSILKITNTE NENENAKKKK RRLNTHEKKL LLDKWADFND SVKKFEAQS SSIENSFVFN FVEGSLVKTI RAGEWLLLDE VNLATADTLE SISDLLTEPD SRSILLSEKG DAEPIKAHPD FRIFACMNPA TDVGKRDLP MGIRSRFTEI YVHSPERDIT DLLSIIDKYI GKYSVSDEWV GNDIAELYLE AKKLSDNNTI VDGSNQKPHF S IRTLTRTL LYVTDIIHIY GLRRSLYDGF CMSFLTLLDQ KSEAILKPVI EKFTLGRLKN VKSIMSQTPP SPGPDYVQFK HY WMKKGPN TIQEQAHYII TPFVEKNMMN LVRATSGKRF PVLIQGPTSS GKTSMIKYLA DITGHKFVRI NNHEHTDLQE YLG TYVTDD TGKLSFKEGV LVEALRKGYW IVLDELNLAP TDVLEALNRL LDDNRELFIP ETQEVVHPHP DFLLFATQNP PGIY GGRKI LSRAFRNRFL ELHFDDIPQD ELEIILRERC QIAPSYAKKI VEVYRQLSIE RSASRLFEQK NSFATLRDLF RWALR DAVG YEQLAASGYM LLAERCRTPQ EKVTVKKTLE KVMKVKLDMD QYYASLEDKS LEAIGSVTWT KGMRRLSVLV SSCLKN KEP VLLVGETGCG KTTICQLLAQ FMGRELITLN AHQNTETGDI LGAQRPVRNR SEIQYKLIKS LKTALNIAND QDVDLKE LL QLYSKSDNKN IAEDVQLEIQ KLRDSLNVLF EWSDGPLIQA MRTGNFFLLD EISLADDSVL ERLNSVLEPE RSLLLAEQ G SSDSLVTASE NFQFFATMNP GGDYGKKELS PALRNRFTEI WVPSMEDFND VNMIVSSRLL EDLKDLANPI VKFSEWFGK KLGGGNATSG VISLRDILAW VEFINKVFPK IQNKSTALIQ GASMVFIDAL GTNNTAYLAE NENDLKSLRT ECIIQLLKLC GDDLELQQI ETNEIIVTQD ELQVGMFKIP RFPDAQSSSF NLTAPTTASN LVRVVRAMQV HKPILLEGSP GVGKTSLITA L ANITGNKL TRINLSEQTD LVDLFGADAP GERSGEFLWH DAPFLRAMKK GEWVLLDEMN LASQSVLEGL NACLDHRGEA YI PELDISF SCHPNFLVFA AQNPQYQGGG RKGLPKSFVN RFSVVFIDML TSDDLLLIAK HLYPSIEPDI IAKMIKLMST LED QVCKRK LWGNSGSPWE FNLRDTLRWL KLLNQYSICE DVDVFDFVDI IVKQRFRTIS DKNKAQLLIE DIFGKFSTKE NFFK LTEDY VQINNEVALR NPHYRYPITQ NLFPLECNVA VYESVLKAIN NNWPLVLVGP SNSGKTETIR FLASILGPRV DVFSM NSDI DSMDILGGYE QVDLTRQISY ITEELTNIVR EIISMNMKLS PNATAIMEGL NLLKYLLNNI VTPEKFQDFR NRFNRF FSH LEGHPLLKTM SMNIEKMTEI ITKEASVKFE WFDGMLVKAV EKGHWLILDN ANLCSPSVLD RLNSLLEIDG SLLINEC SQ EDGQPRVLKP HPNFRLFLTM DPKYGELSRA MRNRGVEIYI DELHSRSTAF DRLTLGFELG ENIDFVSIDD GIKKIKLN E PDMSIPLKHY VPSYLSRPCI FAQVHDILLL SDEEPIEESL AAVIPISHLG EVGKWANNVL NCTEYSEKKI AERLYVFIT FLTDMGVLEK INNLYKPANL KFQKALGLHD KQLTEETVSL TLNEYVLPTV SKYSDKIKSP ESLYLLSSLR LLLNSLNALK LINEKSTHG KIDELTYIEL SAAAFNGRHL KNIPRIPIFC ILYNILTVMS ENLKTESLFC GSNQYQYYWD LLVIVIAALE T AVTKDEAR LRVYKELIDS WIASVKSKSD IEITPFLNIN LEFTDVLQLS RGHSITLLWD IFRKNYPTTS NSWLAFEKLI NL SEKFDKV RLLQFSESYN SIKDLMDVFR LLNDDVLNNK LSEFNLLLSK LEDGINELEL ISNKFLNKRK HYFADEFDNL IRY TFSVDT AELIKELAPA SSLATQKLTK LITNKYNYPP IFDVLWTEKN AKLTSFTSTI FSSQFLEDVV RKSNNLKSFS GNQI KQSIS DAELLLSSTI KCSPNLLKSQ MEYYKNMLLS WLRKVIDIHV GGDCLKLTLK ELCSLIEEKT ASETRVTFAE YIFPA LDLA ESSKSLEELG EAWITFGTGL LLLFVPDSPY DPAIHDYVLY DLFLKTKTFS QNLMKSWRNV RKVISGDEEI FTEKLI NTI SDDDAPQSPR VYRTGMSIDS LFDEWMAFLS STMSSRQIKE LVSSYKCNSD QSDRRLEMLQ QNSAHFLNRL ESGYSKF AD LNDILAGYIY SINFGFDLLK LQKSKDRASF QISPLWSMDP INISCAENVL SAYHELSRFF KKGDMEDTSI EKVLMYFL T LFKFHKRDTN LLEIFEAALY TLYSRWSVRR FRQEQEENEK SNMFKFNDNS DDYEADFRKL FPDYEDTALV TNEKDISSP ENLDDIYFKL ADTYISVFDK DHDANFSSEL KSGAIITTIL SEDLKNTRIE ELKSGSLSAV INTLDAETQS FKNTEVFGNI DFYHDFSIP EFQKAGDIIE TVLKSVLKLL KQWPEHATLK ELYRVSQEFL NYPIKTPLAR QLQKIEQIYT YLAEWEKYAS S EVSLNNTV KLITDLIVSW RKLELRTWKG LFNSEDAKTR KSIGKWWFYL YESIVISNFV SEKKETAPNA TLLVSSLNLF FS KSTLGEF NARLDLVKAF YKHIQLIGLR SSKIAGLLHN TIKFYYQFKP LIDERITNGK KSLEKEIDDI ILLASWKDVN VDA LKQSSR KSHNNLYKIV RKYRDLLNGD AKTIIEAGLL YSNENKLKLP TLKQHFYEDP NLEASKNLVK EISTWSMRAA PLRN IDTVA SNMDSYLEKI SSQEFPNFAD LASDFYAEAE RLRKETPNVY TKENKKRLAY LKTQKSKLLG DALKELRRIG LKVNF REDI QKVQSSTTTI LANIAPFNNE YLNSSDAFFF KILDLLPKLR SAASNPSDDI PVAAIERGMA LAQSLMFSLI TVRHPL SEF TNDYCKINGM MLDLEHFTCL KGDIVHSSLK ANVDNVRLFE KWLPSLLDYA AQTLSVISKY SATSEQQKIL LDAKSTL SS FFVHFNSSRI FDSSFIESYS RFELFINELL KKLENAKETG NAFVFDIIIE WIKANKGGPI KKEQKRGPSV EDVEQAFR R TFTSIILSFQ KVIGDGIESI SETDDNWLSA SFKKVMVNVK LLRSSVVSKN IETALSLLKD FDFTTTESIY VKSVISFTL PVITRYYNAM TVVLERSRIY YTNTSRGMYI LSTILHSLAK NGFCSPQPPS EEVDDKNLQE GTGLGDGEGA QNNNKDVEQD EDLTEDAQN ENKEQQDKDE RDDENEDDAV EMEGDMAGEL EDLSNGEEND DEDTDSEEEE LDEEIDDLNE DDPNAIDDKM W DDKASDNS KEKDTDQNLD GKNQEEDVQA AENDEQQRDN KEGGDEDPNA PEDGDEEIEN DENAEEENDV GEQEDEVKDE EG EDLEANV PEIETLDLPE DMNLDSEHEE SDEDVDMSDG MPDDLNKEEV GNEDEEVKQE SGIESDNEND EPGPEEDAGE TET ALDEEE GAEEDVDMTN DEGKEDEENG PEEQAMSDEE ELKQDAAMEE NKEKGGEQNT EGLDGVEEKA DTEDIDQEAA VQQD SGSKG AGADATDTQE QDDVGGSGTT QNTYEEDQED VTKNNEESRE EATAALKQLG DSMKEYHRRR QDIKEAQTNG EEDEN LEKN NERPDEFEHV EGANTETDTQ ALGSATQDQL QTIDEDMAID DDREEQEVDQ KELVEDADDE KMDIDEEEML SDIDAH DAN NDVDSKKSGF IGKRKSEEDF ENELSNEHFS ADQEDDSEIQ SLIENIEDNP PDASASLTPE RSLEESRELW HKSEIST AD LVSRLGEQLR LILEPTLATK LKGDYKTGKR LNMKRIIPYI ASQFRKDKIW LRRTKPSKRQ YQIMIALDDS KSMSESKC V KLAFDSLCLV SKTLTQLEAG GLSIVKFGEN IKEVHSFDQQ FSNESGARAF QWFGFQETKT DVKKLVAEST KIFERARAM VHNDQWQLEI VISDGICEDH ETIQKLVRRA RENKIMLVFV IIDGITSNES ILDMSQVNYI PDQYGNPQLK ITKYLDTFPF EFYVVVHDI SELPEMLSLI LRQYFTDLAS S UniProtKB: Midasin |
-Macromolecule #2: Ribosome assembly protein 4
| Macromolecule | Name: Ribosome assembly protein 4 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 57.106781 KDa |
| Sequence | String: MSTLIPPPSK KQKKEAQLPR EVAIIPKDLP NVSIKFQALD TGDNVGGALR VPGAISEKQL EELLNQLNGT SDDPVPYTFS CTIQGKKAS DPVKTIDITD NLYSSLIKPG YNSTEDQITL LYTPRAVFKV KPVTRSSSAI AGHGSTILCS AFAPHTSSRM V TGAGDNTA ...String: MSTLIPPPSK KQKKEAQLPR EVAIIPKDLP NVSIKFQALD TGDNVGGALR VPGAISEKQL EELLNQLNGT SDDPVPYTFS CTIQGKKAS DPVKTIDITD NLYSSLIKPG YNSTEDQITL LYTPRAVFKV KPVTRSSSAI AGHGSTILCS AFAPHTSSRM V TGAGDNTA RIWDCDTQTP MHTLKGHYNW VLCVSWSPDG EVIATGSMDN TIRLWDPKSG QCLGDALRGH SKWITSLSWE PI HLVKPGS KPRLASSSKD GTIKIWDTVS RVCQYTMSGH TNSVSCVKWG GQGLLYSGSH DRTVRVWDIN SQGRCINILK SHA HWVNHL SLSTDYALRI GAFDHTGKKP STPEEAQKKA LENYEKICKK NGNSEEMMVT ASDDYTMFLW NPLKSTKPIA RMTG HQKLV NHVAFSPDGR YIVSASFDNS IKLWDGRDGK FISTFRGHVA SVYQVAWSSD CRLLVSCSKD TTLKVWDVRT RKLSV DLPG HKDEVYTVDW SVDGKRVCSG GKDKMVRLWT H UniProtKB: Ribosome assembly protein 4 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R3/3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 0.3 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 20 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: INTEGRATING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Frames/image: 1-48 / Number grids imaged: 1 / Average exposure time: 10.0 sec. / Average electron dose: 75.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT | ||||||||||
| Output model |  PDB-6ylf: |
 Movie
Movie Controller
Controller




















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)