+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0329 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Rea1 AAA2L-H2alpha deletion mutant in AMPPNP State | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Rea1 / Mdn1 / Midasin / AAA+ protein / ribosome maturation / molecular machine / MOTOR PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationprotein-RNA complex remodeling / regulation of ribosomal subunit export from nucleus / preribosome, large subunit precursor / ribosomal large subunit export from nucleus / rRNA processing / ribosomal large subunit assembly / nucleolus / ATP hydrolysis activity / mitochondrion / nucleoplasm ...protein-RNA complex remodeling / regulation of ribosomal subunit export from nucleus / preribosome, large subunit precursor / ribosomal large subunit export from nucleus / rRNA processing / ribosomal large subunit assembly / nucleolus / ATP hydrolysis activity / mitochondrion / nucleoplasm / ATP binding / nucleus Similarity search - Function | |||||||||
| Biological species |  | |||||||||
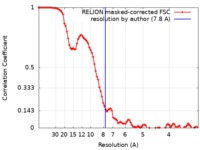
| Method | single particle reconstruction / cryo EM / Resolution: 7.8 Å | |||||||||
 Authors Authors | Sosnowski P / Urnavicius L | |||||||||
| Funding support |  France, 1 items France, 1 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2018 Journal: Elife / Year: 2018Title: The CryoEM structure of the ribosome maturation factor Rea1. Authors: Piotr Sosnowski / Linas Urnavicius / Andreas Boland / Robert Fagiewicz / Johan Busselez / Gabor Papai / Helgo Schmidt /   Abstract: The biogenesis of 60S ribosomal subunits is initiated in the nucleus where rRNAs and proteins form pre-60S particles. These pre-60S particles mature by transiently interacting with various assembly ...The biogenesis of 60S ribosomal subunits is initiated in the nucleus where rRNAs and proteins form pre-60S particles. These pre-60S particles mature by transiently interacting with various assembly factors. The ~5000 amino-acid AAA+ ATPase Rea1 (or Midasin) generates force to mechanically remove assembly factors from pre-60S particles, which promotes their export to the cytosol. Here we present three Rea1 cryoEM structures. We visualise the Rea1 engine, a hexameric ring of AAA+ domains, and identify an α-helical bundle of AAA2 as a major ATPase activity regulator. The α-helical bundle interferes with nucleotide-induced conformational changes that create a docking site for the substrate binding MIDAS domain on the AAA +ring. Furthermore, we reveal the architecture of the Rea1 linker, which is involved in force generation and extends from the AAA+ ring. The data presented here provide insights into the mechanism of one of the most complex ribosome maturation factors. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0329.map.gz emd_0329.map.gz | 5.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0329-v30.xml emd-0329-v30.xml emd-0329.xml emd-0329.xml | 20.5 KB 20.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_0329_fsc.xml emd_0329_fsc.xml | 9.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_0329.png emd_0329.png | 30.4 KB | ||
| Filedesc metadata |  emd-0329.cif.gz emd-0329.cif.gz | 9.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0329 http://ftp.pdbj.org/pub/emdb/structures/EMD-0329 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0329 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0329 | HTTPS FTP |
-Related structure data
| Related structure data |  6i27MC  0308C  0309C  0328C  0330C  6hydC  6hypC  6i26C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_0329.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0329.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.635 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Rea1 (MIDASIN) AAA2L-H2alpha deletion mutant in AMPPNP State - el...
| Entire | Name: Rea1 (MIDASIN) AAA2L-H2alpha deletion mutant in AMPPNP State - electron density map |
|---|---|
| Components |
|
-Supramolecule #1: Rea1 (MIDASIN) AAA2L-H2alpha deletion mutant in AMPPNP State - el...
| Supramolecule | Name: Rea1 (MIDASIN) AAA2L-H2alpha deletion mutant in AMPPNP State - electron density map type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Midasin,Midasin,Midasin,Midasin,Midasin,Midasin,Midasin
| Macromolecule | Name: Midasin,Midasin,Midasin,Midasin,Midasin,Midasin,Midasin type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 546.53825 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) ...String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)PSVPECFTI E KKSSYFII EPQDLSTKVA SICGVIVPKV HTIHDKVFYP LTFVPTHKTV SSLRQLGRKI QNSTPIMLIG KAGSGKTFLI NE LSKYMGC HDSIVKIHLG EQTDAKLLIG TYTSGDKPGT FEWRAGVLAT AVKEGRWVLI EDIDKAPTDV LSILLSLLEK REL TIPSRG ETVKAANGFQ LISTVRINED HQKDSSNKIY NLNMIGMRIW NVIELEEPSE EDLTHILAQK FPILTNLIPK LIDS YKNVK SIYMNTKFIS LNKGAHTRVV SVRDLIKLCE RLDILFKNNG INKPDQLIQS SVYDSIFSEA ADCFAGAIGE FKALE PIIQ AIGESLDIAS SRISLFLTQH VPTLENLDDS IKIGRAVLLK EKLNIQKKSM NSTLFAFTNH SLRLMEQISV CIQMTE PVL LVGETGTGKT TVVQQLAKML AKKLTVINVS QQTETGDLLG GYKPVNSKTV AVPIQENFET LFNATFSLKK NEKFHKM LH RCFNKNQWKN VVKLWNEAYK MAQSILKITN TENENENAKK KKRRLNTHEK KLLLDKWADF NDSVKKFEAQ SSSIENSF V FNFVEGSLVK TIRAGEWLLL DEVNLATADT LESISDLLTE PDSRSILLSE KGDAEPIKAH PDFRIFACMN PATDVGKRD LPMGIRSRFT EIYVHSPERD ITDLLSIIDK YIGKYSVSDE WVGNDIAELY LEAKKLSDNN TIVDGSNQKP HFSIRTLTRT LLYVTDIIH IYGLRRSLYD GFCMSFLTLL DQKSEAILKP VIEKFTLGRL KNVKSIMSQT PPSPGPDYVQ FKHYWMKKGP N TIQEQAHY IITPFVEKNM MNLVRATSGK RFPVLIQGPT SSGKTSMIKY LADITGHKFV RINNHEHTDL QEYLGTYVTD DT GKLSFKE GVLVEALRKG YWIVLDELNL APTDVLEALN RLLDDNRELF IPETQEVVHP HPDFLLFATQ NPPGIYGGRK ILS RAFRNR FLELHFDDIP QDELEIILRE RCQIAPSYAK KIVEVYRQLS IERSASRLFE QKNSFATLRD LFRWALRDAV GYEQ LAASG YMLLAERCRT PQEKVTVKKT LEKVMKVKLD MDQYYASLED KSLEAIGSVT WTKGMRRLSV LVSSCLKNKE PVLLV GETG CGKTTICQLL AQFMGRELIT LNAHQNTETG DILGAQRPVR NRSEIQYKLI KSLKTALNIA NDQDVDLKEL LQLYSK SDN KNIAEDVQLE IQKLRDSLNV LFEWSDGPLI QAMRTGNFFL LDEISLADDS VLERLNSVLE PERSLLLAEQ GSSDSLV TA SENFQFFATM NPGGDYGKKE LSPALRNRFT EIWVPSMEDF NDVNMIVSSR LLEDLKDLAN PIVKFSEWFG KKLGGGNA T SGVISLRDIL AWVEFINKVF PKIQNKSTAL IQGASMVFID ALGTNNTAYL AENENDLKSL RTECIIQLLK LCGDDLELQ QIETNEIIVT QDELQVGMFK IPRFPDAQSS SFNLTAPTTA SNLVRVVRAM QVHKPILLEG SPGVGKTSLI TALANITGNK LTRINLSEQ TDLVDLFGAD APGERSGEFL WHDAPFLRAM KKGEWVLLDE MNLASQSVLE GLNACLDHRG EAYIPELDIS F SCHPNFLV FAAQNPQYQG GGRKGLPKSF VNRFSVVFID MLTSDDLLLI AKHLYPSIEP DIIAKMIKLM STLEDQVCKR KL WGNSGSP WEFNLRDTLR WLKLLNQYSI CEDVDVFDFV DIIVKQRFRT ISDKNKAQLL IEDIFGKFST KENFFKLTED YVQ INNEVA LRNPHYRYPI TQNLFPLECN VAVYESVLKA INNNWPLVLV GPSNSGKTET IRFLASILGP RVDVFSMNSD IDSM DILGG YEQVDLTRQI SYITEELTNI VREIISMNMK LSPNATAIME GLNLLKYLLN NIVTPEKFQD FRNRFNRFFS HLEGH PLLK TMSMNIEKMT EIITKEASVK FEWFDGMLVK AVEKGHWLIL DNANLCSPSV LDRLNSLLEI DGSLLINECS QEDGQP RVL KPHPNFRLFL TMDPKYGELS RAMRNRGVEI YIDELHSRST AFDRLTLGFE LGENIDFVSI DDGIKKIKLN EPDMSIP LK HYVPSYLSRP CIFAQVHDIL LLSDEEPIEE SLAAVIPISH LGEVGKWANN VLNCTEYSEK KIAERLYVFI TFLTDMGV L EKINNLYKPA NLKFQKALGL HDKQLTEETV SLTLNEYVLP TVSKYSDKIK SPESLYLLSS LRLLLNSLNA LKLINEKST HGKIDELTYI ELSAAAFNGR HLKNIPRIPI FCILYNILTV MSENLKTESL FCGSNQYQYY WDLLVIVIAA LETAVTKDEA RLRVYKELI DSWIASVKSK SDIEITPFLN INLEFTDVLQ LSRGHSITLL WDIFRKNYPT TSNSWLAFEK LINLSEKFDK V RLLQFSES YNSIKDLMDV FRLLNDDVLN NKLSEFNLLL SKLEDGINEL ELISNKFLNK RKHYFADEFD NLIRYTFSVD TA ELIKELA PASSLATQKL TKLITNKYNY PPIFDVLWTE KNAKLTSFTS TIFSSQFLED VVRKSNNLKS FSGNQIKQSI SDA ELLLSS TIKCSPNLLK SQMEYYKNML LSWLRKVIDI HVGGDCLKLT LKELCSLIEE KTASETRVTF AEYIFPALDL AESS KSLEE LGEAWITFGT GLLLLFVPDS PYDPAIHDYV LYDLFLKTKT FSQNLMKSWR NVRKVISGDE EIFTEKLINT ISDDD APQS PRVYRTGMSI DSLFDEWMAF LSSTMSSRQI KELVSSYKCN SDQSDRRLEM LQQNSAHFLN RLESGYSKFA DLNDIL AGY IYSINFGFDL LKLQKSKDRA SFQISPLWSM DPINISCAEN VLSAYHELSR FFKKGDMEDT SIEKVLMYFL TLFKFHK RD TNLLEIFEAA LYTLYSRWSV RRFRQEQEEN EKSNMFKFND NSDDYEADFR KLFPDYEDTA LVTNEKDISS PENLDDIY F KLADTYISVF DKDHDANFSS ELKSGAIITT ILSEDLKNTR IEELKSGSLS AVINTLDAET QSFKNTEVFG NIDFYHDFS IPEFQKAGDI IETVLKSVLK LLKQWPEHAT LKELYRVSQE FLNYPIKTPL ARQLQKIEQI YTYLAEWEKY ASSEVSLNNT VKLITDLIV SWRKLELRTW KGLFNSEDAK TRKSIGKWWF YLYESIVISN FVSEKKETAP NATLLVSSLN LFFSKSTLGE F NARLDLVK AFYKHIQLIG LRSSKIAGLL HNTIKFYYQF KPLIDERITN GKKSLEKEID DIILLASWKD VNVDALKQSS RK SHNNLYK IVRKYRDLLN GDAKTIIEAG LLY(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)RNID TVASNMDSYL EKISSQEFPN FADLASDFYA EAERLRKETP NVYTKENKKR L AYLKTQKS KLLGDALKEL RRIGLKVNFR EDIQKVQSST TTILANIAPF NNEYLNSSDA FFFKILDLLP KLRSAASNPS DD IPVAAIE RGMALAQSLM FSLITVRHPL SEFTNDYCKI NGMMLDLEHF TCLKGDIVHS SLKANVDNVR LFEKWLPSLL DYA AQTLSV ISKYSATSEQ QKILLDAKST LSSFFVHFNS SRIFDSSFIE SYSRFELFIN ELLKKLENAK ETGNAFVFDI IIEW IKANK GGPIKKEQKR GPSVEDVEQA FRRTFTSIIL SFQKVIGDGI ESISETDDNW LSASFKKVMV NVKLLRSSVV SKNIE TALS LLKDFDFTTT ESIYVKSVIS FTLPVITRYY NAMTVVLERS RIYYTNTSRG MYILSTILHS LAKNGFCSPQ PPSEEV DDK NLQEGTGLGD GEGAQNNNKD VEQDEDLTED AQNENKEQQD KDERDDENED DAVEMEGDMA GELEDLSNGE ENDDEDT DS EEEELDEEID DLNEDDPNAI DDKMWDDKAS DNSKEKDTDQ NLDGKNQEED VQAAENDEQQ RDNKEGGDED PNAPEDGD E EIENDENAEE ENDVGEQEDE VKDEEGEDLE ANVPEIETLD LPEDMNLDSE HEESDEDVDM SDGMPDDLNK EEVGNEDEE VKQESGIESD NENDEPGPEE DAGETETALD EEEGAEEDVD MTNDEGKEDE ENGPEEQAMS DEEELKQDAA MEENKEKGGE QNTEGLDGV EEKADTEDID QEAAVQQDSG SKGAGADATD TQEQDDVGGS GTTQNTYEED QEDVTKNNEE SREEATAALK Q LGDSMKEY HRRRQDIKEA QTNGEEDENL EKNNERPDEF EHVEGANTET DTQALGSATQ DQLQTIDEDM AIDDDREEQE VD QKELVED ADDEKMDIDE EEMLSDIDAH DANNDVDSKK SGFIGKRKSE EDFENELSNE HFSADQEDDS EIQSLIENIE DNP PDASAS LTPERSLEES RELWHKSEIS TADLVSRLGE QLRLILEPTL ATKLKGDYKT GKRLNMKRII PYIASQFRKD KIWL RRTKP SKRQYQIMIA LDDSKSMSES KCVKLAFDSL CLVSKTLTQL EAGGLSIVKF GENIKEVHSF DQQFSNESGA RAFQW FGFQ ETKTDVKKLV AESTKIFERA RAMVHNDQWQ LEIVISDGIC EDHETIQKLV RRARENKIML VFVIIDGITS NESILD MSQ VNYIPDQYGN PQLKITKYLD TFPFEFYVVV HDISELPEML SLILRQYFTD LASS UniProtKB: Midasin, Midasin |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.2 Component:
Details: AMP-PNP was added 5 minute before the plunging | ||||||||||||||||||
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 12 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 280 K / Instrument: FEI VITROBOT MARK III |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Phase plate: VOLTA PHASE PLATE / Spherical aberration corrector: Titan Krios Cs Corrector / Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3712 pixel / Digitization - Dimensions - Height: 3840 pixel / Digitization - Frames/image: 2-36 / Number grids imaged: 1 / Number real images: 2479 / Average exposure time: 0.2 sec. / Average electron dose: 46.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 105000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 0.01 mm / Nominal defocus max: 0.6 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)