+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0308 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Rea1 Wild type ADP state (tail part) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Rea1 / Mdn1 / Midasin / AAA+ protein / ribosome maturation / molecular machine / MOTOR PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationprotein-RNA complex remodeling / regulation of ribosomal subunit export from nucleus / preribosome, large subunit precursor / ribosomal large subunit export from nucleus / rRNA processing / ribosomal large subunit assembly / nucleolus / ATP hydrolysis activity / mitochondrion / nucleoplasm ...protein-RNA complex remodeling / regulation of ribosomal subunit export from nucleus / preribosome, large subunit precursor / ribosomal large subunit export from nucleus / rRNA processing / ribosomal large subunit assembly / nucleolus / ATP hydrolysis activity / mitochondrion / nucleoplasm / ATP binding / nucleus Similarity search - Function | |||||||||
| Biological species |   | |||||||||
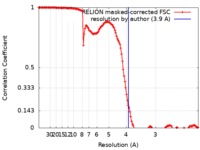
| Method | single particle reconstruction / cryo EM / Resolution: 3.9 Å | |||||||||
 Authors Authors | Sosnowski P / Urnavicius L | |||||||||
| Funding support |  France, 1 items France, 1 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2018 Journal: Elife / Year: 2018Title: The CryoEM structure of the ribosome maturation factor Rea1. Authors: Piotr Sosnowski / Linas Urnavicius / Andreas Boland / Robert Fagiewicz / Johan Busselez / Gabor Papai / Helgo Schmidt /   Abstract: The biogenesis of 60S ribosomal subunits is initiated in the nucleus where rRNAs and proteins form pre-60S particles. These pre-60S particles mature by transiently interacting with various assembly ...The biogenesis of 60S ribosomal subunits is initiated in the nucleus where rRNAs and proteins form pre-60S particles. These pre-60S particles mature by transiently interacting with various assembly factors. The ~5000 amino-acid AAA+ ATPase Rea1 (or Midasin) generates force to mechanically remove assembly factors from pre-60S particles, which promotes their export to the cytosol. Here we present three Rea1 cryoEM structures. We visualise the Rea1 engine, a hexameric ring of AAA+ domains, and identify an α-helical bundle of AAA2 as a major ATPase activity regulator. The α-helical bundle interferes with nucleotide-induced conformational changes that create a docking site for the substrate binding MIDAS domain on the AAA +ring. Furthermore, we reveal the architecture of the Rea1 linker, which is involved in force generation and extends from the AAA+ ring. The data presented here provide insights into the mechanism of one of the most complex ribosome maturation factors. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0308.map.gz emd_0308.map.gz | 11.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0308-v30.xml emd-0308-v30.xml emd-0308.xml emd-0308.xml | 19.9 KB 19.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_0308_fsc.xml emd_0308_fsc.xml | 13.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_0308.png emd_0308.png | 36.4 KB | ||
| Filedesc metadata |  emd-0308.cif.gz emd-0308.cif.gz | 7.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0308 http://ftp.pdbj.org/pub/emdb/structures/EMD-0308 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0308 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0308 | HTTPS FTP |
-Related structure data
| Related structure data |  6hydMC  0309C  0328C  0329C  0330C  6hypC  6i26C  6i27C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_0308.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0308.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Rea1 (MIDASIN) tail with ADP
| Entire | Name: Rea1 (MIDASIN) tail with ADP |
|---|---|
| Components |
|
-Supramolecule #1: Rea1 (MIDASIN) tail with ADP
| Supramolecule | Name: Rea1 (MIDASIN) tail with ADP / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Midasin,Midasin,Midasin
| Macromolecule | Name: Midasin,Midasin,Midasin / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 192.045828 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: PIEESLAAVI PISHLGEVGK WANNVLNCTE YSEKKIAERL YVFITFLTDM GVLEKINNLY KPANLKFQKA LGLHDKQLTE ETVSLTLNE YVLPTVSKYS DKIKSPESLY LLSSLRLLLN SLNALKLINE KSTHGKIDEL TYIELSAAAF NGRHLKNIPR I PIFCILYN ...String: PIEESLAAVI PISHLGEVGK WANNVLNCTE YSEKKIAERL YVFITFLTDM GVLEKINNLY KPANLKFQKA LGLHDKQLTE ETVSLTLNE YVLPTVSKYS DKIKSPESLY LLSSLRLLLN SLNALKLINE KSTHGKIDEL TYIELSAAAF NGRHLKNIPR I PIFCILYN ILTVMSENLK TESLFCGSNQ YQYYWDLLVI VIAALETAVT KDEARLRVYK ELIDSWIASV KSKSDIEITP FL NINLEFT DVLQLSRGHS ITLLWDIFRK NYPTTSNSWL AFEKLINLSE KFDKVRLLQF SESYNSIKDL MDVFRLLNDD VLN NKLSEF NLLLSKLEDG INELELISNK FLNKRKHYFA DEFDNLIRYT FSVDTAELIK ELAPASSLAT QKLTKLITNK YNYP PIFDV LWTEKNAKLT SFTSTIFSSQ FLEDVVRKSN NLKSFSGNQI KQSISDAELL LSSTIKCSPN LLKSQMEYYK NMLLS WLRK VIDIHVGGDC LKLTLKELCS LIEEKTASET RVTFAEYIFP ALDLAESSKS LEELGEAWIT FGTGLLLLFV PDSPYD PAI HDYVLYDLFL KTKTFSQNLM KSWRNVRKVI SGDEEIFTEK LINTISDDDA PQSPRVYRTG MSIDSLFDEW MAFLSST MS SRQIKELVSS YKCNSDQSDR RLEMLQQNSA HFLNRLESGY SKFADLNDIL AGYIYSINFG FDLLKLQKSK DRASFQIS P LWSMDPINIS CAENVLSAYH ELSRFFKKGD MEDTSIEKVL MYFLTLFKFH KRDTNLLEIF EAALYTLYSR WSVRRFRQE QEENEKSNMF KFNDNSDDYE ADFRKLFPDY EDTALVTNEK DISSPENLDD IYFKLADTYI SVFDKDHDAN FSSELKSGAI ITTILSEDL KNTRIEELKS GSLSAVINTL DAETQSFKNT EVFGNIDFYH DFSIPEFQKA GDIIETVLKS VLKLLKQWPE H ATLKELYR VSQEFLNYPI KTPLARQLQK IEQIYTYLAE WEKYASSEVS LNNTVKLITD LIVSWRKLEL RTWKGLFNSE DA KTRKSIG KWWFYLYESI VISNFVSEKK ETAPNATLLV SSLNLFFSKS TLGEFNARLD LVKAFYKHIQ LIGLRSSKIA GLL HNTIKF YYQFKPLIDE RITNGKKSLE KEIDDIILLA SWKDVNVDAL KQSSRKSHNN LYKIVRKYRD LLNGDAKTII EAGL LY(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) RNIDTVASNM DSYLEKISSQ EFPNFADLAS DFYAEAERLR KETPNVYTKE NKKRLAYLKT QKSKLLGDAL KELRRIGLKV NFREDIQKV QSSTTTILAN IAPFNNEYLN SSDAFFFKIL DLLPKLRSAA SNPSDDIPVA AIERGMALAQ SLMFSLITVR H PLSEFTND YCKINGMMLD LEHFTCLKGD IVHSSLKANV DNVRLFEKWL PSLLDYAAQT LSVISKYSAT SEQQKILLDA KS TLSSFFV HFNSSRIFDS SFIESYSRFE LFINELLKKL ENAKETGNAF VFDIIIEWIK ANKGGPIKKE QKRGPSVEDV EQA FRRTFT SIILSFQKVI GDGIESISET DDNWLSASFK KVMVNVKLLR SSVVSKNIET ALSLLKDFDF TTTESIYVKS VISF TLPVI TRYYNAMTVV LERSRIYYTN TSRGMYILST ILHSLAKN UniProtKB: Midasin, Midasin |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.2 Component:
Details: ADP was added 5 minute before the plunging | ||||||||||||||||||
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 12 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Instrument: HOMEMADE PLUNGER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Spherical aberration corrector: Titan Krios Cs Corrector / Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3712 pixel / Digitization - Dimensions - Height: 3840 pixel / Digitization - Frames/image: 2-35 / Number grids imaged: 14 / Number real images: 23230 / Average exposure time: 0.2 sec. / Average electron dose: 46.2 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 105000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 0.01 mm / Nominal defocus max: 3.4 µm / Nominal defocus min: 1.8 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)