[English] 日本語
 Yorodumi
Yorodumi- EMDB-10836: Rix1-Rea1 pre-60S particle - Rix1-subcomplex, body 3 (rigid body ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10836 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Rix1-Rea1 pre-60S particle - Rix1-subcomplex, body 3 (rigid body refinement) | |||||||||
 Map data Map data | Rix1-subcomplex of the Rix1-Rea1 pre-60S particle | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | pre-60S / Biogenesis / LSU / Large subunit / ribosome assembly / RIBOSOME | |||||||||
| Function / homology |  Function and homology information Function and homology informationrixosome complex / pre-replicative complex assembly involved in nuclear cell cycle DNA replication / nuclear pre-replicative complex / regulation of DNA-templated DNA replication initiation / Major pathway of rRNA processing in the nucleolus and cytosol / maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / DNA-templated DNA replication / rRNA processing / ribosomal large subunit assembly / chromatin binding ...rixosome complex / pre-replicative complex assembly involved in nuclear cell cycle DNA replication / nuclear pre-replicative complex / regulation of DNA-templated DNA replication initiation / Major pathway of rRNA processing in the nucleolus and cytosol / maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / DNA-templated DNA replication / rRNA processing / ribosomal large subunit assembly / chromatin binding / nucleoplasm / nucleus / cytosol Similarity search - Function | |||||||||
| Biological species |  | |||||||||
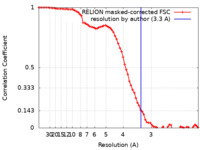
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Kater L / Beckmann R | |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2020 Journal: Mol Cell / Year: 2020Title: Construction of the Central Protuberance and L1 Stalk during 60S Subunit Biogenesis. Authors: Lukas Kater / Valentin Mitterer / Matthias Thoms / Jingdong Cheng / Otto Berninghausen / Roland Beckmann / Ed Hurt /  Abstract: Ribosome assembly is driven by numerous assembly factors, including the Rix1 complex and the AAA ATPase Rea1. These two assembly factors catalyze 60S maturation at two distinct states, triggering ...Ribosome assembly is driven by numerous assembly factors, including the Rix1 complex and the AAA ATPase Rea1. These two assembly factors catalyze 60S maturation at two distinct states, triggering poorly understood large-scale structural transitions that we analyzed by cryo-electron microscopy. Two nuclear pre-60S intermediates were discovered that represent previously unknown states after Rea1-mediated removal of the Ytm1-Erb1 complex and reveal how the L1 stalk develops from a pre-mature nucleolar to a mature-like nucleoplasmic state. A later pre-60S intermediate shows how the central protuberance arises, assisted by the nearby Rix1-Rea1 machinery, which was solved in its pre-ribosomal context to molecular resolution. This revealed a Rix1-Ipi3 tetramer anchored to the pre-60S via Ipi1, strategically positioned to monitor this decisive remodeling. These results are consistent with a general underlying principle that temporarily stabilized immature RNA domains are successively remodeled by assembly factors, thereby ensuring failsafe assembly progression. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10836.map.gz emd_10836.map.gz | 4.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10836-v30.xml emd-10836-v30.xml emd-10836.xml emd-10836.xml | 19.1 KB 19.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10836_fsc.xml emd_10836_fsc.xml | 7.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_10836.png emd_10836.png | 59.8 KB | ||
| Filedesc metadata |  emd-10836.cif.gz emd-10836.cif.gz | 6.4 KB | ||
| Others |  emd_10836_half_map_1.map.gz emd_10836_half_map_1.map.gz emd_10836_half_map_2.map.gz emd_10836_half_map_2.map.gz | 28.2 MB 28.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10836 http://ftp.pdbj.org/pub/emdb/structures/EMD-10836 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10836 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10836 | HTTPS FTP |
-Related structure data
| Related structure data |  6yleMC  6ylfC  6ylgC  6ylhC  6ylxC  6ylyC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10836.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10836.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Rix1-subcomplex of the Rix1-Rea1 pre-60S particle | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.059 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: Relion half map (unfiltered, unmasked)
| File | emd_10836_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Relion half map (unfiltered, unmasked) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Relion half map (unfiltered, unmasked)
| File | emd_10836_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Relion half map (unfiltered, unmasked) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Rix1-Rea1 pre-60S assembly particle - Rix1-subcomplex
| Entire | Name: Rix1-Rea1 pre-60S assembly particle - Rix1-subcomplex |
|---|---|
| Components |
|
-Supramolecule #1: Rix1-Rea1 pre-60S assembly particle - Rix1-subcomplex
| Supramolecule | Name: Rix1-Rea1 pre-60S assembly particle - Rix1-subcomplex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Rix1-TAP Flag-Rea1 derived pre-60S assembly complex |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Pre-rRNA-processing protein IPI3
| Macromolecule | Name: Pre-rRNA-processing protein IPI3 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 61.836695 KDa |
| Sequence | String: MDEQVIFTTN TSGTIASVHS FEQINLRQCS TQSRNSCVQV GNKYLFIAQA QKALINVYNL SGSFKRESVE QRLPLPEILK CLEVVENDG VQYDRIQGVN HNLPDFNLPY LLLGSTESGK LYIWELNSGI LLNVKPMAHY QSITKIKSIL NGKYIITSGN D SRVIIWQT ...String: MDEQVIFTTN TSGTIASVHS FEQINLRQCS TQSRNSCVQV GNKYLFIAQA QKALINVYNL SGSFKRESVE QRLPLPEILK CLEVVENDG VQYDRIQGVN HNLPDFNLPY LLLGSTESGK LYIWELNSGI LLNVKPMAHY QSITKIKSIL NGKYIITSGN D SRVIIWQT VDLVSASNDD PKPLCILHDH TLPVTDFQVS SSQGKFLSCT DTKLFTVSQD ATIRCYDLSL IGSKKKQKAN EN DVSIGKT PVLLATFTTP YSIKSIVLDP ADRACYIGTA EGCFSLNLFY KLKGNAIVNL LQSAGVNTVQ KGRVFSLVQR NSL TGGENE DLDALYAMGQ LVCENVLNSN VSCLEISMDG TLLLIGDTEG KVSIAEIYSK QIIRTIQTLT TSQDSVGEVT NLLT NPYRL ERGNLLFEGE SKGKQPSNNN GHNFMKIPNL QRVIFDGKNK GHLHDIWYQI GEPEAETDPN LALPLNDFNA YLEQV KTQE SIFSHIGKVS SNVKVIDNKI DATSSLDSNA AKDEEITELK TNIEALTHAY KELRDMHEKL YEEHQQMLDK Q UniProtKB: Pre-rRNA-processing protein IPI3 |
-Macromolecule #2: Pre-rRNA-processing protein RIX1
| Macromolecule | Name: Pre-rRNA-processing protein RIX1 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 86.839844 KDa |
| Sequence | String: MSEEFIAVST LARNLEIAKG NEFHTILATL RSPVYINEQL LKSELSFLVT KILKLIRSGN DFDLWKGCHT SVVTCAYNPL VLSTHGGQL LAAIYSRLEQ KTGFYSSVIS SSHGKQLFNT LISSVAIIID LMKNKPTLSR EALVPKLKAI IPTLITLSQY E PELVLPVL ...String: MSEEFIAVST LARNLEIAKG NEFHTILATL RSPVYINEQL LKSELSFLVT KILKLIRSGN DFDLWKGCHT SVVTCAYNPL VLSTHGGQL LAAIYSRLEQ KTGFYSSVIS SSHGKQLFNT LISSVAIIID LMKNKPTLSR EALVPKLKAI IPTLITLSQY E PELVLPVL QRILKRNTTT FKPFTNKFRT VLINLIISDY ASLGTKTQRL VCENFAYLHL LKIQVSDTSD DETQAHHKIY AD SNWRTGL MSILSQFKPI IQLCGEILDF EQDNELYKLI KSLPVIDESN NKEEFLPSLK LDFNAPLTLW EIPQRLSLLA DML VAFISL PTPFPIRVPL GGINSLCEVL LGVSNKYLPL KKELRHDNEL NGVINTILPQ IQFQGIRLWE IMVSKYGKCG LSFF EGILS SIELFIPLKK KSNNEIDFNV VGSLKFEFAT VFRLVNMILS HLGHQLNIIS VISQLIEVAL FLSHDKTLID SLFKN RKSI MKQQTKTKQS KRSKSAEGAF SDIYTHPELF VCKNSMNWFN EINDFFITAL NNWILPSTPH IQILKYSITQ SLRLKE RFG YIPESFVNLL RCEVLHPGSE RVSILPIAIS LLKNINDDMF ELLCHPKVPV GMVYQLHKPL DLGEDGEVRD DINKKEV ET NESSSNANTG LETLKALENL ENVTIPEPKH EVPKVVDDTA IFKKRSVEEV IERESTSSHK KVKFVEETTV DNGEELIV K KAVSQTKEEE KPMEDSEDEE QEEFEIPAIE LSDDEEEEEE EE UniProtKB: Pre-rRNA-processing protein RIX1 |
-Macromolecule #3: Pre-rRNA-processing protein IPI1
| Macromolecule | Name: Pre-rRNA-processing protein IPI1 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 37.928895 KDa |
| Sequence | String: MTKSRKQKQK KQDFLRKKLK VGKPKEKARN ATDTSFVSKT ISIRNQHLDQ NPHDLTKRLT LLKHHNINVR KETLTTFQKS IPSIIKSRL MTPLLTQSIP LICDESQQVR QGLIDLVDEI GSHDAEILKL HCNIFVLYIN MAMTHIVTQI QADSTKFLSH L LKYCGDEV ...String: MTKSRKQKQK KQDFLRKKLK VGKPKEKARN ATDTSFVSKT ISIRNQHLDQ NPHDLTKRLT LLKHHNINVR KETLTTFQKS IPSIIKSRL MTPLLTQSIP LICDESQQVR QGLIDLVDEI GSHDAEILKL HCNIFVLYIN MAMTHIVTQI QADSTKFLSH L LKYCGDEV VRKSWVKLLN GVFGVLGWGQ VGKNDSASIV QTKKRNAKYV TIHLNALYTL VEYGCQDERA RSDGDTAETT ED SGTLRNP YLIPDYPQPF EHLKLFTREL KVQDATSSGV NATLLSLATQ DIDTRKAVFI EQFLPIVRKK IEVIIKEGGE CGK SANKLK TLLAKIFD UniProtKB: Pre-rRNA-processing protein IPI1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R3/3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 20 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: INTEGRATING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Frames/image: 1-48 / Number grids imaged: 1 / Average exposure time: 10.0 sec. / Average electron dose: 75.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-6yle: |
 Movie
Movie Controller
Controller



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)