[English] 日本語
 Yorodumi
Yorodumi- EMDB-0977: Cyanobacterial PSI Monomer from T. elongatus by Single Particle C... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0977 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cyanobacterial PSI Monomer from T. elongatus by Single Particle CRYO-EM at 3.2 A Resolution | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | monomer / complex / photosystem / photosynthesis / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationphotosystem I reaction center / photosystem I / photosynthetic electron transport in photosystem I / photosystem I / plasma membrane-derived thylakoid membrane / chlorophyll binding / photosynthesis / 4 iron, 4 sulfur cluster binding / electron transfer activity / oxidoreductase activity ...photosystem I reaction center / photosystem I / photosynthetic electron transport in photosystem I / photosystem I / plasma membrane-derived thylakoid membrane / chlorophyll binding / photosynthesis / 4 iron, 4 sulfur cluster binding / electron transfer activity / oxidoreductase activity / magnesium ion binding / metal ion binding Similarity search - Function | |||||||||
| Biological species |   Thermosynechococcus elongatus BP-1 (bacteria) / Thermosynechococcus elongatus BP-1 (bacteria) /   Thermosynechococcus elongatus (strain BP-1) (bacteria) Thermosynechococcus elongatus (strain BP-1) (bacteria) | |||||||||
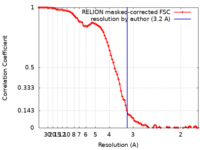
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Kurisu G / Coruh O | |||||||||
| Funding support |  Japan, 2 items Japan, 2 items
| |||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2021 Journal: Commun Biol / Year: 2021Title: Cryo-EM structure of a functional monomeric Photosystem I from Thermosynechococcus elongatus reveals red chlorophyll cluster. Authors: Orkun Çoruh / Anna Frank / Hideaki Tanaka / Akihiro Kawamoto / Eithar El-Mohsnawy / Takayuki Kato / Keiichi Namba / Christoph Gerle / Marc M Nowaczyk / Genji Kurisu /    Abstract: A high-resolution structure of trimeric cyanobacterial Photosystem I (PSI) from Thermosynechococcus elongatus was reported as the first atomic model of PSI almost 20 years ago. However, the monomeric ...A high-resolution structure of trimeric cyanobacterial Photosystem I (PSI) from Thermosynechococcus elongatus was reported as the first atomic model of PSI almost 20 years ago. However, the monomeric PSI structure has not yet been reported despite long-standing interest in its structure and extensive spectroscopic characterization of the loss of red chlorophylls upon monomerization. Here, we describe the structure of monomeric PSI from Thermosynechococcus elongatus BP-1. Comparison with the trimer structure gave detailed insights into monomerization-induced changes in both the central trimerization domain and the peripheral regions of the complex. Monomerization-induced loss of red chlorophylls is assigned to a cluster of chlorophylls adjacent to PsaX. Based on our findings, we propose a role of PsaX in the stabilization of red chlorophylls and that lipids of the surrounding membrane present a major source of thermal energy for uphill excitation energy transfer from red chlorophylls to P700. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0977.map.gz emd_0977.map.gz | 59.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0977-v30.xml emd-0977-v30.xml emd-0977.xml emd-0977.xml | 34.4 KB 34.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_0977_fsc.xml emd_0977_fsc.xml | 9.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_0977.png emd_0977.png | 296.5 KB | ||
| Filedesc metadata |  emd-0977.cif.gz emd-0977.cif.gz | 8.3 KB | ||
| Others |  emd_0977_half_map_1.map.gz emd_0977_half_map_1.map.gz emd_0977_half_map_2.map.gz emd_0977_half_map_2.map.gz | 49.6 MB 49.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0977 http://ftp.pdbj.org/pub/emdb/structures/EMD-0977 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0977 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0977 | HTTPS FTP |
-Related structure data
| Related structure data |  6lu1MC  7bw2C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10352 (Title: 3.2 Å resolution structure of a functional monomeric Photosystem I from Thermosynechococcus elongatus BP-1 by single particle cryo-EM with a 200 kV CRYO ARM electron microscope EMPIAR-10352 (Title: 3.2 Å resolution structure of a functional monomeric Photosystem I from Thermosynechococcus elongatus BP-1 by single particle cryo-EM with a 200 kV CRYO ARM electron microscopeData size: 226.1 Data #1: Raw, non aligned micrograph movies in compressed TIFF format. [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_0977.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0977.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.89 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: #1
| File | emd_0977_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_0977_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : Cyanobacterial PSI Monomer from T. elongatus
+Supramolecule #1: Cyanobacterial PSI Monomer from T. elongatus
+Macromolecule #1: Photosystem I P700 chlorophyll a apoprotein A1
+Macromolecule #2: Photosystem I P700 chlorophyll a apoprotein A2
+Macromolecule #3: Photosystem I iron-sulfur center
+Macromolecule #4: Photosystem I reaction center subunit II
+Macromolecule #5: Photosystem I reaction center subunit IV
+Macromolecule #6: Photosystem I reaction center subunit III
+Macromolecule #7: Photosystem I reaction center subunit VIII
+Macromolecule #8: Photosystem I reaction center subunit IX
+Macromolecule #9: Photosystem I reaction center subunit XI
+Macromolecule #10: Photosystem I reaction center subunit XII
+Macromolecule #11: CHLOROPHYLL A
+Macromolecule #12: PHYLLOQUINONE
+Macromolecule #13: BETA-CAROTENE
+Macromolecule #14: 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE
+Macromolecule #15: IRON/SULFUR CLUSTER
+Macromolecule #16: 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE
+Macromolecule #17: water
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 20 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV | ||||||||||||
| Details | Sample was monodisperse. |
- Electron microscopy
Electron microscopy
| Microscope | JEOL CRYO ARM 200 |
|---|---|
| Temperature | Min: 100.4 K / Max: 100.4 K |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3710 pixel / Digitization - Dimensions - Height: 3838 pixel / Digitization - Frames/image: 1-60 / Number grids imaged: 1 / Average exposure time: 12.0 sec. / Average electron dose: 1.34 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated defocus max: 3.5 µm / Calibrated defocus min: 0.5 µm / Calibrated magnification: 56497 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 1.4 mm / Nominal magnification: 60000 |
| Sample stage | Specimen holder model: JEOL / Cooling holder cryogen: NITROGEN |
 Movie
Movie Controller
Controller



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)