+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6822 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of atOSCA1.1 channel | ||||||||||||
 Map data Map data | sharpened, used for model building | ||||||||||||
 Sample Sample |
| ||||||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of calcium ion import / mechanosensitive monoatomic ion channel activity / cellular hyperosmotic response / calcium-activated cation channel activity / response to osmotic stress / monoatomic cation channel activity / protein tetramerization / plasma membrane / cytosol Similarity search - Function | ||||||||||||
| Biological species |  | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.52 Å | ||||||||||||
 Authors Authors | Chen L / Zhang M / Kang Y / Wu JX | ||||||||||||
| Funding support |  China, 3 items China, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2018 Journal: Nat Struct Mol Biol / Year: 2018Title: Structure of the mechanosensitive OSCA channels. Authors: Mingfeng Zhang / Dali Wang / Yunlu Kang / Jing-Xiang Wu / Fuqiang Yao / Chengfang Pan / Zhiqiang Yan / Chen Song / Lei Chen /  Abstract: Mechanosensitive ion channels convert mechanical stimuli into a flow of ions. These channels are widely distributed from bacteria to higher plants and humans, and are involved in many crucial ...Mechanosensitive ion channels convert mechanical stimuli into a flow of ions. These channels are widely distributed from bacteria to higher plants and humans, and are involved in many crucial physiological processes. Here we show that two members of the OSCA protein family in Arabidopsis thaliana, namely AtOSCA1.1 and AtOSCA3.1, belong to a new class of mechanosensitive ion channels. We solve the structure of the AtOSCA1.1 channel at 3.5-Å resolution and AtOSCA3.1 at 4.8-Å resolution by cryo-electron microscopy. OSCA channels are symmetric dimers that are mediated by cytosolic inter-subunit interactions. Strikingly, they have structural similarity to the mammalian TMEM16 family proteins. Our structural analysis accompanied with electrophysiological studies identifies the ion permeation pathway within each subunit and suggests a conformational change model for activation. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6822.map.gz emd_6822.map.gz | 59.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6822-v30.xml emd-6822-v30.xml emd-6822.xml emd-6822.xml | 16.3 KB 16.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_6822.png emd_6822.png | 223.3 KB | ||
| Masks |  emd_6822_msk_1.map emd_6822_msk_1.map | 64 MB |  Mask map Mask map | |
| Others |  emd_6822_half_map_1.map.gz emd_6822_half_map_1.map.gz emd_6822_half_map_2.map.gz emd_6822_half_map_2.map.gz | 59.2 MB 59.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6822 http://ftp.pdbj.org/pub/emdb/structures/EMD-6822 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6822 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6822 | HTTPS FTP |
-Related structure data
| Related structure data |  6jpfMC  6875C  5z1fC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_6822.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6822.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened, used for model building | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
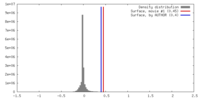
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_6822_msk_1.map emd_6822_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
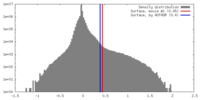
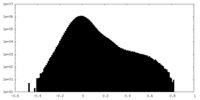
| Density Histograms |
-Half map: #1
| File | emd_6822_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
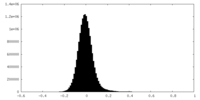
| Density Histograms |
-Half map: #2
| File | emd_6822_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : atOSCA1.1
| Entire | Name: atOSCA1.1 |
|---|---|
| Components |
|
-Supramolecule #1: atOSCA1.1
| Supramolecule | Name: atOSCA1.1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 175 KDa |
-Macromolecule #1: Protein OSCA1
| Macromolecule | Name: Protein OSCA1 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 87.697008 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MATLKDIGVS AGINILTAFI FFIIFAFLRL QPFNDRVYFS KWYLRGLRSS PASGGGFAGR FVNLELRSYL KFLHWMPEAL KMPERELID HAGLDSVVYL RIYWLGLKIF APIAMLAWAV LVPVNWTNNE LELAKHFKNV TSSDIDKLTI SNIPEGSNRF W AHIIMAYA ...String: MATLKDIGVS AGINILTAFI FFIIFAFLRL QPFNDRVYFS KWYLRGLRSS PASGGGFAGR FVNLELRSYL KFLHWMPEAL KMPERELID HAGLDSVVYL RIYWLGLKIF APIAMLAWAV LVPVNWTNNE LELAKHFKNV TSSDIDKLTI SNIPEGSNRF W AHIIMAYA FTIWTCYMLM KEYETVANMR LQFLASEGRR PDQFTVLVRN VPPDPDETVS ELVEHFFLVN HPDNYLTHQV VC NANKLAD LVSKKTKLQN WLDYYQLKYT RNNSQIRPIT KLGCLGLCGQ KVDAIEHYIA EVDKTSKEIA EERENVVNDQ KSV MPASFV SFKTRWAAAV CAQTTQTRNP TEWLTEWAAE PRDIYWPNLA IPYVSLTVRR LVMNVAFFFL TFFFIIPIAF VQSL ATIEG IEKVAPFLKV IIEKDFIKSL IQGLLAGIAL KLFLIFLPAI LMTMSKFEGF TSVSFLERRS ASRYYIFNLV NVFLG SVIA GAAFEQLNSF LNQSPNQIPK TIGMAIPMKA TFFITYIMVD GWAGVAGEIL MLKPLIIYHL KNAFLVKTEK DREEAM NPG SIGFNTGEPQ IQLYFLLGLV YAPVTPMLLP FILVFFALAY VVYRHQIINV YNQEYESAAA FWPDVHGRVI TALIISQ LL LMGLLGTKHA ASAAPFLIAL PVITIGFHRF CKGRFEPAFV RYPLQEAMMK DTLERAREPN LNLKGYLQDA YIHPVFKG G DNDDDGDMIG KLENEVIIVP TKRQSRRNTP APSRISGESS PSLAVINGKE V |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 0.01 mm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-6jpf: |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)